Genetic Testing for Epilepsy - CAM 266
Description
Epilepsy is a group of disorders characterized by recurrent, unprovoked seizures due to abnormal, synchronized neuronal firing in the brain that can be distinguished by seizure type, age of onset, developmental status, comorbid features, and etiology.1,2
For guidance on the use of chromosomal microarray or low-pass whole genome sequencing for the diagnosis of epilepsy, please reference policy CAM 313 Chromosomal Microarray and Low-pass Whole Genome Sequencing.
Regulatory Status
Many labs have developed specific tests that they must validate and perform in house. These laboratory-developed tests (LDTs) are regulated by the Centers for Medicare and Medicaid (CMS) as high-complexity tests under the Clinical Laboratory Improvement Amendments of 1988 (CLIA ’88). LDTs are not approved or cleared by the U. S. Food and Drug Administration; however, FDA clearance or approval is not currently required for clinical use.
Policy
Application of coverage criteria is dependent upon an individual’s benefit coverage at the time of the request.
- Genetic counseling IS REQUIRED for individuals prior to and after undergoing genetic testing for epilepsy and when whole exome sequencing (WES) or whole genome sequencing (WGS) is being considered, counseling must be provided by an ABMGG board-certified medical geneticist or an ABGC board-certified genetic counselor.
- For individuals with an epilepsy syndrome (in which epilepsy is the core clinical symptom), WES or multi-gene panel testing for variants associated with epilepsy syndromes (see Note 1) is considered MEDICALLY NECESSARY only when a positive test result would lead to changes in at least one of the following:
- Medication management.
- Diagnostic testing (alternative potentially invasive tests are avoided).
- Reproductive decision making.
- When WES is unable to identify a causative variant and the clinical suspicion of a genomic etiology remains in situations where any of the above criteria are met in their entirety, WGS is considered MEDICALLY NECESSARY.
- For individuals with an epilepsy syndrome with a suspected genetic cause, targeted variant testing is considered MEDICALLY NECESSARY to assess for the following clinical situations:
- ALDH7A1 testing for pyridoxine-related epilepsy.
- BTD testing for biotinidase deficiency-related epilepsy.
- FOLR1 testing for cerebral folate deficiency-related epilepsy.
- GRIN2A testing for GRIN2A-related epileptic encephalopathy.
- KCNQ2 testing for KCNQ2-related epileptic encephalopathy.
- KCNT1 testing for KCNT1-related migrating partial epilepsy of infancy.
- PCDH19 testing for epilepsy female-restricted with mental retardation (EFMR).
- PHGDH, PSAT1, and PSPH testing for epilepsy due to serine biosynthesis defects.
- SCN1A testing for SCN1A-related seizure disorders.
- SCN8A testing for SCN8A-related epileptic encephalopathy.
- SLC2A1 testing for glucose transporter type 1 deficiency syndrome.
- SLC6A8, GATM, and GAMT testing for epilepsy due to creatine deficiency syndromes.
- TSC1 and TSC2 testing for tuberous sclerosis complex-related epilepsy.
The following does not meet coverage criteria due to a lack of available published scientific literature confirming that the test(s) is/are required and beneficial for the diagnosis and treatment of a patient’s illness.
- For all other situations and/or mutations not described above, genetic testing for epilepsy investigational/unproven therefore considered NOT MEDICALLY NECESSARY.
NOTES:
Note 1: For two or more gene tests being run on the same platform, please refer to CAM 235 Reimbursement Policy. Panel testing for epilepsy must target 25 or more genes associated with epilepsy.
Policy Guidelines
Coding
If the specific gene being tested has been codified in CPT, the appropriate CPT code would be reported. If the specific gene has not been codified in CPT, the unlisted molecular pathology code 81479 would be reported. If a panel of tests that has not been codified in CPT is performed, code 81479 would be reported once.
Here is a list of some of the tests related to epilepsy that are listed under CPT tier 2 codes:
Under CPT code 81401:
MT-TK (mitochondrially encoded tRNA lysine) (e.g., myoclonic epilepsy with ragged-red fibers [MERRF]), common variants (e.g., m.8344A>G, m.8356T>C)
Under CPT code 81403:
NHLRC1 (NHL repeat containing 1) (e.g., progressive myoclonus epilepsy), full gene sequence
Under CPT code 81404:
ARX (aristaless related homeobox) (e.g., X-linked lissencephaly with ambiguous genitalia, X-linked mental retardation), full gene sequence
EPM2A (epilepsy, progressive myoclonus type 2A, Lafora disease [laforin]) (e.g., progressive myoclonus epilepsy), full gene sequence.
Under CPT code 81405:
CHRNA4 (cholinergic receptor, nicotinic, alpha 4) (e.g., nocturnal frontal lobe epilepsy), full gene sequence.
CHRNB2 (cholinergic receptor, nicotinic, beta 2 [neuronal]) (e.g., nocturnal frontal lobe epilepsy), full gene sequence
GABRG2 (gamma-aminobutyric acid [GABA] A receptor, gamma 2) (e.g., generalized epilepsy with febrile seizures), full gene sequence.
Under CPT code 81406:
ALDH7A1 (aldehyde dehydrogenase 7 family, member A1) (e.g., pyridoxine-dependent epilepsy), full gene sequence
CDKL5 (cyclin-dependent kinase-like 5) (e.g., early infantile epileptic encephalopathy), full gene sequence
EFHC1 (EF-hand domain (C-terminal) containing 1) (e.g., juvenile myoclonic epilepsy), full gene sequence
Under CPT code 81407:
SCN1A (sodium channel, voltage-gated, type 1, alpha subunit) (e.g., generalized epilepsy with epilepsy with febrile seizures), full gene sequence.
Table of Terminology
| Term |
Definition |
| AAN |
American Academy of Neurology |
| ALDH7A1 |
Aldehyde dehydrogenase 7 family member A1 |
| AMA |
Antiseizure medication |
| ANK2 |
Ankyrin 2 |
| ASD |
Autism spectrum disorders |
| BTD |
Biotinidase |
| CACNA1A |
Calcium voltage-gated channel subunit alpha 1A |
| CACNA1AE |
Calcium voltage-gated channel subunit alpha 1AE |
| CACNA1D |
Calcium voltage-gated channel subunit alpha 1D |
| CACNA1H |
Calcium voltage-gated channel subunit alpha 1H |
| CACNA2D3 |
Calcium voltage-gated channel subunit alpha 1A |
| CDKL5 |
Calcium voltage-gated channel subunit alpha2delta3 |
| CGH |
Comparative genomic hybridization |
| CHD2 |
Chromodomain helicase DNA binding protein 2 |
| CLIA ’88 |
Clinical Laboratory Improvement Amendments of 1988 |
| CMA |
Chromosomal microarray analysis |
| CMS |
Centers for Medicare & Medicaid Services |
| CNS |
Child Neurology Society |
| CNV |
Copy number variant |
| DEE |
Developmental and epileptic encephalopathy |
| DLG4 |
Discs large homolog 4 |
| DS |
Dravet syndrome |
| EE |
Epileptic encephalopathy |
| EEG |
Electroencephalography |
| EFMR |
Epilepsy female-restricted with mental retardation |
| EFNS |
European Federation of Neurological Societies |
| EP |
Epileptic panels |
| FGF12 |
Fibroblast growth factor 12 |
| FOLR1 |
Folate receptor 1 |
| GABRA1 |
Gamma-aminobutyric acid type A receptor alpha1 |
| GABRG2 |
Gamma-aminobutyric acid type A receptor gamma2 |
| GAMT |
Guanidinoacetate n-methyltransferase |
| GATM |
Glycine amidinotransferase |
| GGE |
Genetic generalized epilepsy |
| Glut1DS |
Glucose transporter 1 deficiency syndrome |
| GRIA2 |
Glutamate ionotropic receptor AMPA type subunit 2 |
| GRIN2A |
Glutamate ionotropic receptor NMDA type subunit 2a |
| GRIN2B |
Glutamate ionotropic receptor NMDA type subunit 2b |
| GS |
Genome sequencing |
| GTAC |
Genetic Testing Advisory Committee |
| HNRNPU |
Heterogeneous nuclear ribonucleoprotein U |
| IC-CODE |
International Classification of Cognitive Disorders in Epilepsy |
| ID |
Intellectual disability |
| ILAE |
International League Against Epilepsy |
| KANSL1 |
KAT8 regulatory NSL complex subunit 1 |
| KB |
Kilo-base pair |
| KCNC1 |
Potassium voltage-gated channel subfamily C member 1 |
| KCNQ2 |
Potassium voltage-gated channel subfamily Q member 2 |
| KCNT1 |
Potassium sodium-activated channel subfamily T member 1 |
| KD |
Ketogenic diet |
| LDTs |
Laboratory-developed tests |
| MECP2 |
Methyl-CpG binding protein 2 |
| MGP |
Multigene panel |
| mTOR |
Mammalian target of rapamycin |
| NDD |
Neurodevelopmental disorders |
| NGS |
Next-generation sequencing |
| NICE |
National Institute for Health and Care Excellence |
| PCDH19 |
Protocadherin 19 |
| PHGDH |
Phosphoglycerate dehydrogenase |
| PNPO |
Pyridoxamine 5'-phosphate oxidase |
| PRRT2 |
Proline-rich transmembrane protein 2 |
| PSAT1 |
Phosphoserine aminotransferase 1 |
| PSPH |
Phosphoserine phosphatase |
| PURA |
Pur-alpha |
| SCN1A |
Sodium voltage-gated channel alpha subunit 1 |
| SCN2A |
Sodium voltage-gated channel type 2 alpha subunit |
| SCN8A |
Sodium voltage-gated channel type 8 alpha subunit |
| SE |
Status epilepticus |
| SIGN |
Scottish Intercollegiate Guidelines Network |
| SLC2A1 |
Solute carrier family 2 member 1 |
| SLC6A1 |
Solute carrier family 6 member 1 |
| SLC6A8 |
Solute carrier family 6 member 8 |
| SMEI |
Severe myoclonic epilepsy of infancy |
| STX1B |
Syntaxin 1B |
| STXBP1 |
Syntaxin-binding protein 1 |
| TCF4 |
Transcription factor 4 |
| TSC |
Tuberous sclerosis complex |
| TSC1 |
Tuberous sclerosis complex 1 |
| TSC2 |
Tuberous sclerosis complex 2 |
| UBE3A |
Ubiquitin protein ligase E3A |
| WES |
Whole exome sequencing |
| WGS |
Whole genome sequencing |
| YWHAG |
Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein gamma |
Rationale
EEpilepsy, defined as having two or more unprovoked seizures, is a common neurologic disorder, affecting an estimated three million adults and 470,000 children in the United States.3 The biological and genetic mechanisms that disturb the balance between excitatory and inhibitory neuronal circuits to result in epilepsy are extremely heterogeneous.4,5 Approximately 20–30% of epilepsy diagnoses can be attributed to other primary conditions, such as stroke, tumor or head injury, but the remaining 70–80% of cases are believed to be due to one or more genetic factors.2,6
The epilepsies can be classified by multiple approaches. Clinically, they can be broadly grouped into three classes: genetic generalized epilepsy (GGE), focal epilepsy, and epileptic encephalopathy (EE). GGE is characterized by generalized seizures that involve both sides of the brain, start in childhood or adolescence, are usually associated with normal development and intellect, and include juvenile myoclonic epilepsy and childhood absence epilepsy, among others. Focal seizures originate in one hemisphere of the brain and include temporal lobe epilepsy and autosomal dominant nocturnal frontal lobe epilepsy. EE are severe, early-onset conditions characterized by refractory seizures, developmental delay or regression associated with ongoing epileptic activity, and generally poor prognosis such as Dravet, Ohtahara, and West syndromes. Epilepsy is often a comorbid condition with intellectual disability (ID), autism, or schizophrenia, and may be a feature of many metabolic conditions and genetic syndromes.2
The International League Against Epilepsy (ILAE) classifies epilepsies at three levels: seizure types, epilepsy types, and epilepsy syndromes. The classification starts with seizure type as the least specific, epilepsy type as more specific, and epilepsy syndromes as the most specific classification. Seizure type is based on onset, of which there are three categories: focal, generalized, and unknown. The second category, epilepsy type, is divided into four categories; focal, generalized, combined generalized & focal, and unknown. This level of classification typically requires electroencephalography (EEG) data. The “combined generalized & focal” group is new to the 2017 edition of the ILAE classification, and it was created to include patients with both types of seizures, such as patients with Dravet syndrome. Finally, the “Unknown” category refers to a case where there is not enough information to determine the epilepsy type. This can occur for numerous reasons, such as the unavailability of an EEG. The third level of classification is “epilepsy syndromes.” A syndrome refers to a cluster of features incorporating items such as seizure types, EEG results, and imaging results that tend to occur together. Other distinctive features, such as ID, may be a part of a syndrome.7
Some conditions have well-known mutations caused by a singular gene. For example, in Dravet syndrome, (DS, previously known as severe myoclonic epilepsy of infancy or SMEI) the occurrence of SCN1A mutations is 70%-80%. About 90% of these mutations are de novo. Truncating mutations have found to be more severe (earlier onset of symptoms, faster cognitive decline) compared to missense mutations. PCDH19-related epilepsy is a rare syndrome characterized by focal and/or generalized seizures, which are commonly fever-induced and in clusters. Previously referred to as EFMR, PCDH19-related epilepsy occurs primarily in females and has an early-onset. A mutation in PCDH19 can cause both DS and PCDH19-related epilepsy as PCDH19 is thought to account for 5% of patients with DS.8 Another example is tuberous sclerosis complex (TSC); as many as 89% of cases have mutations in either the TSC1 or TSC2 gene.9-11 Other mutations include KCNQ2 with EE, SLC2A1 with glucose transporter deficiency syndrome, PRRT2 with general infantile convulsions, and ALDH7A1 and PNPO with severe early-onset epilepsy.12 Missense variants in SLC32A1 can cause genetic epilepsy with febrile seizures plus (GEFS+) and idiopathic generalizes epilepsy (IGE).13
Epilepsy is genetically heterogeneous, and extensive phenotypic heterogeneity has been observed even in many monogenic epilepsies. This heterogeneity makes testing each gene individually through Sanger sequencing impractical despite its status as the gold standard.14 Mutations in ion channels, chromatin remodeling, transcriptional regulation, and regulation of the mammalian target of rapamycin (mTOR) protein have been implicated in the etiology of epilepsy.15
Clinical Utility and Validity
The DNA sequencing methods for epilepsy diagnosis have high analytic validity; the methods are well-established in laboratories. Sequencing bi-directionally has been evaluated at over 98% sensitivity.16 Next-generation sequencing (NGS) allows for parallel sequencing of any number of genes, thereby allowing for far superior testing of heterogenous conditions such as epilepsy. Since the number of new epilepsy genes continues to grow and the phenotypes associated with mutations in each gene are so variable, NGS is a desirable option to diagnose epilepsy patients.17
New test options for epilepsy diagnosis have an increased yield of molecular diagnosis, particularly in patients with severe, early-onset epilepsies. For example, a study by Lemke, et al. (2012) focused on targeted resequencing of 265 candidate genes, which identified mutations that were presumed to be pathogenic in 16 of 33 patients. Many of these patients had severe epilepsies associated with ID. This NGS method detected mutations that had been missed by the previous gold standard of Sanger sequencing (with at least one mutation missed due to artifacts from Sanger sequencing). Furthermore, the authors concluded that a patient with more than one possible gene responsible for their phenotype would benefit from this sequencing method compared to Sanger. The authors noted that NGS is only suited for conditions with at least one major gene effect and that WES would be a better option for more complex genetic conditions.14
In 2016, a gene panel targeting 46 epilepsy genes was used on 216 patients representing a wide spectrum of epilepsies with age of onset spanning from the neonatal period to adulthood. A presumed pathogenic variant was identified in 49 (23%) patients. The variants were found in 19 different genes including SCN1A, STXBP1, and CDKL5. Patients with neonatal-onset epilepsies had the highest rate of positive findings (57%). The overall yield for patients with EEs was 32%, compared to 17% among patients with generalized epilepsies and 16% in patients with focal or multifocal.15 Another study focusing on the diagnostic use of microarray was performed by Hrabik, et al. (2015). Approximately 17.7% (26/147) of participants between the ages of birth to 23 years who were diagnosed with epilepsy and had a SNP microarray performed had an abnormal microarray as defined by laboratory guidelines.18 Gene panels and exome sequencing for clinical diagnostics provide better and less expensive options for testing and could be implemented early in the diagnostic process. Chromosome microarrays may be considered in severe cases and in GGE with comorbid features.2 Overall, sequencing of many genes at once may provide major benefits to those with epilepsy as epilepsy’s genetic profile is widespread.
Copy number abnormalities also play an important role in patients with epilepsy. Of 973 patients who had chromosomal microarray (CMA) and ICD-9 codes for epilepsy or seizures, 805 patients satisfied criteria for epilepsy. There were 437 copy number variants (CNVs) in 323 patients (1-4 per patient), including 185 (42%) deletions and 252 (58%) duplications were observed. Forty (9%) were confirmed de novo, 186 (43%) were inherited, and parental data were unavailable for 211 (48%). Because the diagnostic yield of CMA for epilepsy patients is similar to the yield in autism spectrum disorders (ASD) and in prenatal diagnosis (which published guidelines recommending testing with CMA), the authors recommended implementation of CMA in the evaluation of unexplained epilepsy.19
Berg, et al. (2017) conducted a prospective cohort study of 775 children with newly diagnosed epilepsy with an onset at less than three years of age to determine the role of genetic testing in the initial evaluation of early-life epilepsies. Ninety-five children had brain injury, and of the other 680, 327 underwent genetic testing, such as microarrays, WES, and gene panels. In total, 132 (40.4%) children were found to have pathogenic variants. A total of 446 children were deemed to have an unknown etiology without genetic testing, and 180 of these children were tested. Pathogenic variants were found in 48 or 26.7% of the children, however, epilepsy gene-sequencing panels and WES had substantially greater diagnostic yields than CMA. Epilepsy panels detected 28 of 96 (29.2%), WES detected five of 18 or (27.8%), and CMA detected eight for 101 or (7.9%). Without a clinically identified cause, testing yields were greater than 15% and as high as 47% depending on patient subgroups. The authors concluded that broad genetic sequencing methods have high diagnostic yields in diagnosing early-life epilepsies regardless of clinical features. Sequencing tests should be incorporated into the initial evaluation of newly presenting early-life epilepsies and not just reserved for those with severe presentations.20
Gene panels including the most mutated genes have a 10%-50% diagnostic rate (depending on the panel used and the population) although it is possible that a greater number of genes in the panel does not necessarily equate to a higher diagnostic rate. A 67 gene panel and a 265 gene panel were found to have nearly equivalent diagnostic rates; however, the study using the 67 gene panel only included nineteen patients.17,21 Exome sequencing has been evaluated at a 25% diagnostic rate (without a prior diagnosis), and WGS has been evaluated at as high as 60% with smaller studies and known phenotypes. The weaker sequencing depth contributes to the difficulty of locating CNVs, which are a significant cause of epilepsy-related conditions. Nonetheless, the authors conclude that WES and WGS will become part of the regularly used tools for clinicians once their cost decreases.21
Stosser, et al. (2017) conducted a retrospective analysis of “893 probands with epilepsy who had an epilepsy panel or WES performed and were positive for a pathogenic or likely pathogenic variant in one of nine genes (CDKL5, GABRA1, GABRG2, GRIN2B, KCNQ2, MECP2, PCDH19, SCN1A, or SCN2A),” which found “mosaic pathogenic variants… at an overall frequency of 3.5% (95% CI, 2.4%-4.9%) in nine genes associated with epilepsy-related disorders…. Mosaicism was most common in the CDKL5, PCDH19, SCN2A, and SCN1A genes. Mosaicism was observed in GABRA1, GABRG2, and GRIN2B, which were not reported to have mosaicism prior to this study, and in KCNQ2 and MECP2. Parental mosaicism was observed for pathogenic variants in multiple genes including KCNQ2, MECP2, SCN1A, and SCN2A.” The authors concluded that patients with epilepsy who previously tested negative for pathogenic variants may benefit from an NGS test, which is superior at detecting mosaic variants. The authors also noted that targeted testing of parents of probands may use NGS to better assess risk and mosaicism.22
“Epilepsy-plus” patients tend to have highest yield for diagnostic testing; that is, patients with other symptoms such as autism in addition to their epilepsy. Still, even the diagnostic yield of these individuals only reached 50% with panel or exome sequencing.23 The potential clinical utility of genetic testing for these syndromes is in avoiding further diagnostic testing, directing medication management, and assisting in reproductive decision making. Establishing the genetic basis of epilepsy in a given patient will help in making treatment decisions, genetic counseling, and more.12 For instance, mutations in ALDH7A1 or in the PNPO gene may lead to seizures not treated with typical antiepileptic drugs, but with pyridoxine.24 On the other hand, sodium channel agents may be avoided in patients with an SCNA1 mutation. Genetic counseling revolving around topics such as recurrence or heritability risk for pregnancies or testing of relatives is also helpful, and proper care must be taken to ensure that the information is clearly explained to the patient and their family.12
Genetic testing for epilepsy could also be useful in diagnosing ASD. Peng, et al. (2021) discussed, in their research on developing a multiplex gene and phenotype network to identify shared genes between epilepsy and autism, that mutations in genes for subunits of ion channels were related to epilepsy, and that general ion channel dysfunctions “are also linked to susceptibility to autism, as well as bipolar disorder, schizophrenia, and other neuropsychiatric disorders.” The researchers “prioritize ANK2, CACNA1AE, CACNA2D3, GRIA2, and DLG4… as candidate epilepsy genes because of their overlap with the epilepsy-focused module 2 of the WES [whole-exome sequencing] network;” these genes originated as having association only with autism prior to this study.25 A study with similar intention done on co-occurring epilepsy and ASD in Chinese children found SCN1A and MECP2 gene mutations to be the most common; SCN1A was more associated with epilepsy, while MECP2 was more associated with Rett Syndrome. Mutations in SCN2A, CACNA1A, CACNA1H, CACNA1D, and KCNQ2 were also identified, although individual cases for each gene were small in the latter set of genes.26 Both studies contribute to the understanding of the beneficial utility for genetic testing of epilepsy in the context of other comorbidities, such as ASD.
Sanchez Fernandez, et al. (2019) evaluated the cost-effectiveness of genetic testing in patients with epilepsy of unknown etiology. There were 20 studies included, with eight evaluating chromosomal microarray (CMA), nine evaluating epileptic panels (EP) with deletion/duplication testing, and six evaluating WES. The authors found WES to have the highest diagnostic yield at 0.45, followed by EP at 0.23 and then CMA at 0.08. EP was found to be the most cost-effective, at $15,848 per diagnosis. Although cost-effectiveness of strategies overlapped, the authors found CMA to be consistently less cost-effective than WES and EP.27
Borlot, et al. (2019) analyzed the results of epilepsy gene panels from 64 patients. Up to 185 genes were tested. Fourteen probands were found to have “pathogenic or likely pathogenic” variants in the following genes: “SCN1A, GABRB3, UBE3A, KANSL1, SLC2A1, KCNQ2, SLC6A1, HNRNPU, STX1B, SCN2A, PURA, and CHD2”. The authors also identified six mutations arising de novo, with unknown inheritance for eight mutations. Overall, the authors concluded that a commercial gene panel for epilepsy may be useful, as it detected etiology in 22% of patients with epilepsy and ID.28
Kim, et al. (2021) analyzed the clinical utility of WES for patients with infantile-onset epilepsy that had tested negative on gene panel tests for epilepsy. The study included 59 patients. Following WES, 55.4% of the participants received genetic conformation of epilepsy, indicating that WES increased diagnostic yield by 8%. Three epilepsy-causing genes (that were not included on the original gene panel) were identified: YWHAG, KCNC1, and FGF12. The authors conclude that WES could be an important way to reanalyze novel epilepsy-linked genes without updated gene panels.29
Willimsky, et al. (2021) studied the use of NGS by comping the diagnostic yield of small and large gene panels. The authors completed a retrospective study of 190 patients under the age of 18 years who were diagnosed with epilepsy of unknown etiology. Small gene panels were defined as those under 25 kilo-base pair (kb), and large gene panels were defined as those over 25 kb. Diagnostic yield was defined as detection of pathogenic or likely pathogenic variants. The authors found that the diagnostic yield of large panels (29%) was significantly larger than small panels (13%). The authors then analyzed the differences in diagnostic yield in developmental and epileptic encephalography (DEE) and non-DEE. The authors found that the significant increase of diagnostic yield in large panels was only significant for non-DEE patients, and not for DEE patients. The authors conclude that large epilepsy gene panels have significantly higher diagnostic yield for non-DEE patients but note that small gene panels (a maximum of ten genes) is sufficient for DEE patients.30
Stefanski, et al. (2021) completed a meta-analysis and systematic review on the success rate of genetic testing of neurodevelopmental disorders (NDD). The study measured the diagnostic yield, defined as the “percent of pathogenic variant carriers identified in a cohort.” The study included 103 clinical sequencing studies that used NGS in a total of 32,331 people with epilepsy, ASD, or ID. The diagnostic yield for epilepsy was 24%. The epilepsy subtypes with the highest diagnostic yield were epilepsy with ID (27.9%) and early-onset seizures (36.9%). The authors then studied the diagnostic yield of each sequencing technology and found a diagnostic yield of 27.2% for exome sequencing and a diagnostic yield of 22.6% for target gene-sequencing panels.31
Sheidley, et al. (2021) completed a similar meta-analysis and systematic review to Stefanski, et al. (2021), except on 5985 studies on genetic testing of patients with epilepsy. “The overall diagnostic yield across all test modalities was 17%, with the highest yield for GS [genome sequencing] (48%), followed by ES [exome sequencing] (24%), MGP [multigene panel] (19%), and [genome-wide comparative genomic hybridization/chromosomal microarray] CGH/CMA (9%).” The authors also studied non-yield outcomes. A total of 24 studies reported on changes in treatment based on genetic testing; “treatment changes were reported in 12-18% of patients with a genetic diagnosis, including avoiding, stopping, or initiating specific antiseizure medications (AMAs) or ketogenic diet (KD) and halting a plan for surgery in the presence of a specific genetic diagnosis.”32
Beyond developing panels to identify genetic variants among epileptic patients, recent advancements in the study of epigenetics could potentially be of use with epilepsy symptoms but not a clear-cut genetic basis. In the patients with neurodevelopmental disorders an epilepsy phenotype, 23% of cases had rare differential methylations.33 “When the parents were able to be tested, ~40% of the methylation variants were de novo, suggesting that methylation abnormalities may be causative in 5-10% of their cohort. When identified, the underlying causes of the methylation changes were varied and included CNVs (copy number variants), sequence variants in regularly elements, or repeat expansions, each of which is easily missed by conventional (even next-generation) sequencing methods.”34 Genetic and epigenetic testing could further help shape precision medicine in the best types of pharmaceutical drugs used to treat specific epilepsies, such as by understanding individual responses to antiepileptic drug treatment and alterations in drug pharmacokinetics and pharmacodynamics that could guide safer and more effective treatment strategies.35
McKnight, et al. (2022) conducted a study on the diagnostic yield of genetic tests performed on adults with epilepsy. The study included 2,008 individuals and the authors analyzed their NGS targeted testing results of 89-189 genes and clinical information. Diagnostic yield was defined as a definitive molecular diagnosis. In total, 218 adults (10.9%) had clinically actionable findings. The authors compared the diagnostic yield with age of seizure onset and found that “the highest diagnostic yield was in adults with seizure onset during infancy (29.6%, 0–1 year), followed by in early childhood (13.6%, 2–4 years), late childhood (7.0%, 5–10 years), adolescence (2.4%, 11–17 years), and adulthood (3.7%, ≥18 years).” The authors also reported that “comorbid intellectual disability (ID) or developmental delay resulted in a high diagnostic yield (16.0%).” Overall, the authors conclude that genetic testing could impact clinical management and outcomes.36
International League Against Epilepsy (ILAE)
In 2015, the ILAE Commission of Pediatrics issued a task force report which included the following related to genetic testing in epilepsy.
- “Genetic screening should not be undertaken at a primary or secondary level of care.”
- “Standard care should permit genetic counseling by trained personnel to be undertaken at all levels of care (primary to quaternary).”
- “Genetic evaluation for Dravet syndrome and other infantile-onset epileptic encephalopathies should be available at tertiary and quaternary levels of care (optimal intervention would permit an extended genetic evaluation).”
- “Early diagnosis of some mitochondrial conditions may alter long-term outcome, but whether screening at quaternary level is beneficial is unknown.”37
In 2017, the ILAE released a position paper containing a stance on the genetic etiologies of epilepsies. The ILAE states that “the epilepsies in which a genetic etiology has been implicated are quite diverse and, in most cases, the underlying genes are not yet known”. The ILAE also notes that genetic is not equivalent to inherited; de novo mutations are being identified with increasing frequency, thereby making an epileptic syndrome not inherited. Furthermore, the ILAE states that environmental contributions to an epileptic syndrome should not be minimized and that a genetic etiology should be of significant effect in causing epilepsy.7
In the 2021 report, the ILAE and the International Neuropsychological Society address the lack of an international taxonomy for cognitive disorders in epilepsy and proposes the International Classification of Cognitive Disorders in Epilepsy (IC-CODE), stating that “a taxonomy built on the alignment of genetic, neuroimaging, and cognitive processes promises high precision for the diagnosis of cognitive disorders in epilepsy and their treatment.”38
In 2022, ILAE released a position statement on epilepsy syndromes with onset in neonates and infants. The statement notes that “genetic testing [for Dravet syndrome, previously known as severe myoclonic epilepsy of infancy] is recommended at all ages, including in adults in whom the diagnosis is suspected but details of history in infancy may be difficult to access. A pathogenic variant in SCN1A is present in more than 80%–85% of cases.” The statement did not mention recommendations on genetic testing for any other form of epilepsy.39
In 2022, the ILAE released a position statement on idiopathic generalized epilepsy syndromes. Genetic testing is mentioned throughout the statement in relation to epilepsy occurring in individuals with intellectual disabilities. In patients suspected of childhood absence epilepsy or generalized tonic-clonic seizures alone, “investigations, including genetic testing, to exclude other etiologies should be considered” in individuals with intellectual disabilities. All other mentions of genetic testing for the diagnosis of epilepsy note that genetic testing is not currently part of the routine diagnostic evaluation but may be used soon, as more genetic determinants are identified.40
European Academy of Neurology (EAN)
In 2010, the European Federation of Neurological Societies (EFNS) issued the following recommendations pertaining to epilepsy. The EFNS noted that molecular investigations may be useful for diagnosis but cannot be considered routine with the large number of mutations present and state that clinical or physiological methods are generally superior. However, they note SMEI has a mutation in the gene SCN1A in 80% of patients. Since 2010, this organization has been renamed to the European Academy of Neurology (EAN), but no new updates on epilepsy have been released.41
In 2024 the EAN released a statement on the importance of genetic testing in adults with epilepsy, including the use of large gene panels and the role of genetic counseling. “[G]ene panels with ≤511 genes miss a significant number of causative genes in adult patients with epilepsy and intellectual disability”, which is why they recommend that “either large gene panels or exome sequencing should be used to increase the diagnostic yield in this cohort.”42
American Academy of Neurology (AAN) and Child Neurology Society (CNS)
The AAN and CNS issued guidelines for clinicians on diagnostic assessment of the child with status epilepticus (SE) in 2006, reaffirmed 2019. The recommendations provided guidance for the assessment of laboratory studies, metabolic and genetic studies, electroencephalography, and neuroimaging in children with SE. The expert panel concluded that “there are insufficient data to support or refute whether genetic testing (chromosomal or molecular studies) should be done routinely in children with SE.”43
American Epilepsy Society Annual Meeting
In 2015, a talk presented at the American Epilepsy Society Annual Meeting focused on reviewing the genetics of epilepsy, which groups were likely to benefit from genetic testing, and how to approach genetic testing in the current evolution of medicine. Regarding whom would benefit most, “it would make sense to focus testing at this time on those in whom there is a high likelihood of a diagnostic finding and on those whose refractory epilepsy may be influenced by a precise genetic diagnosis that can guide treatment.” This study concluded that genetic testing is needed to determine if treatment regimens will be affected, especially in younger children and infants.
With regards to the proper way to approach genetic testing, “we should start with a clinical approach, defining a specific epilepsy syndrome when relevant, to guide the nature and sequence of genetic testing. Pre- and post- testing counseling, whether provided by a physician or genetic counselor, is critical to maintaining clear communication with patients about the implications and limitations of their genetic testing.” The types of testing include chromosomal microarray analysis (CMA), complete GS, and exome sequencing.23
In 2023, the AES reviewed the 2022 NSGC guideline in a press release, emphasizing genetic testing for adults with epilepsy. They highlighted that over 40% of adult epilepsy cases have a genetic link. Despite genetic testing being more common in children, many adults experience delays up to 10 years after their first seizure before testing. The AES underscores that early testing can lead to significant changes in care, such as adjustments to antiseizure medication, surgical decisions, and family planning guidance. The AES has endorsed the NSGC’s guidelines, which recommend genetic testing for all patients with unexplained epilepsy, regardless of age.44
National Society of Genetic Counselors (NSGC)
In 2022, the NSGC, supported by the American Epilepsy Society, released a clinical guideline on genetic testing and counseling for unexplained epilepsies that provides evidence-based recommendations to guide healthcare providers in diagnostic and management strategies. These guidelines emphasize the importance of genetic testing and counseling as integral parts of care for individuals with unexplained epilepsy. The NSGC guideline includes the following evidence-based recommendations:
- "Genetic testing is strongly recommended for all individuals with unexplained epilepsy, without limitation of age, with exome/genome sequencing and/or a multi-gene panel (>25 genes) as first-tier testing followed by chromosomal microarray, with exome/genome sequencing conditionally recommended over multi-gene panel.”45
- “It is strongly recommended that genetic tests be selected, ordered, and interpreted by a qualified healthcare provider in the setting of appropriate pre-test and post-test genetic counseling.”45
International Glut1DS Study Group
The International Glut1DS (glucose transporter 1 deficiency syndrome) put forth recommendations on the diagnosis and treatment of Glut1DS in 2020, which involves “characteristic clinical features, definite hypoglycorrhachia, and pathogenic SLC2A1 variants.” Hypoglycorrhachia is defined as a low glucose level in cerebrospinal fluid in the setting normal glucose levels in blood, or normoglycemia. Their strength of support for a combination of the


three features is shown below.
On genetic testing, the International Glut1DS group also stated that “the absence of SLC2A1 pathogenic variants does not always exclude Glut1DS…SLC2A1-negative patients can be diagnosed on the basis of hypoglycorrhachia and distinctive clinical features, especially when responsive to KDT [ketogenic diet therapies].”46
North American Consensus Panel on Dravet Syndrome
This panel was convened to establish standards for evaluation of DS. The panel recommended that genetic testing be performed in patients whose symptoms indicated DS; the panel noted that either specific SCN1A sequencing or a larger gene panel may be performed, and a chromosomal microarray was not necessary.
The panel also notes that genetic testing should be pursued in a child <12 months and with ≥2 febrile seizures >15 minutes as well as a child 12-35 months old with at least one febrile seizure over 15 minutes. However, genetic testing should not be performed for a child <12 months with only one focal or generalized febrile seizure.
Genetic counseling should be provided, particularly information about heritability and risk of epilepsy in siblings. Other information about SCN1A mutations may also be provided.47
Dravet Syndrome Foundation
The Dravet syndrome Foundation released a 2022 diagnosis and treatment guideline for clinical presentation, genetic testing, treatments for seizures and other aspects of DS. Recommendations include when and how genetic testing should be conducted for individuals suspected of having DS.
- “SCN1A mutations are found in over 85% of patients clinically diagnosed with Dravet syndrome”. Testing for SCN1A mutations is recommended for patients who meet the clinical testing criteria outlined below.
- “Rarely, dominant pathogenic variants in other genes have been associated with Dravet syndrome, including GABRG2, GABRA1, and STXBP1, and rare recessive cases of SCN1B variants.”48
Genetic Testing Criteria:
“Experts strongly agree that genetic testing should be considered for all patients who meet any one of the following criteria, assuming the child’s early development is normal, there are no causal lesions, and the seizure etiology remains unknown:
- Single prolonged (5-29 min) hemiclonic seizure or focal/generalized status epilepticus (> 29 min) in the context of vaccination or fever in a child aged 2-15 months.
- Single prolonged generalized tonic-clonic seizure in the context of vaccination or fever in a child aged 2-5 months.
- Single prolonged generalized convulsive seizure in the context of vaccination in a child aged 6-15 months.
- Single episode of afebrile convulsive status epilepticus in a child aged 2-15 months.
- Single, prolonged afebrile hemiclonic seizure in a child aged 2-15 months.
- Recurrent prolonged focal or generalized convulsive seizures with or without fever in a child aged 2-15 months.
- Recurrent brief hemiclonic seizures without fever in a child aged 6-15 months.
- Recurrent, hemiclonic seizures with or without fever in child aged 2-15 months.
- Genetic testing is recommended in patients of ALL ages, including in adults with a suspected diagnosis but for whom a detailed history of presentation in infancy may be limited.
- While simple SCN1A sequencing is appropriate when all clinical criteria are met, an epilepsy gene panel is the expert preference for children with suspected DS.”48
Scottish Intercollegiate Guidelines Network (SIGN)
The SIGN lists features suggesting genetic generalized epilepsies, which are as follows:
- childhood or teenage onset
- triggered by sleep deprivation and alcohol
- early morning tonic-clonic seizures or myoclonic jerks
- short absence seizures
- photo paroxysmal response on electroencephalography (EEG)
- generalized three per second spike and wave or polyspike and wave on EEG.
Genetics services are recommended for patients with a “very strong” family history of epilepsy, or with “a clinical phenotype suggestive of a monogenic epilepsy syndrome.”49
National Institute for Health and Care Excellence (NICE)
The NICE clinical guideline on epilepsies, in children, young people, and adults was updated in 2022 to include recommendations on genetic testing.
- Discuss with a neurologist or geneticist any uncertainties about whether to offer genetic testing or which tests to offer to a person with epilepsy.
- Before carrying out genetic tests:
- discuss the purpose of testing and the possible implications of the results with the person with epilepsy, and their family and carers if appropriate
- obtain informed consent with appropriate genetic counseling in line with the NHS Genomic Medicine Service.
- Consider WGS for people with epilepsy of unknown cause who:
- were aged under two years when epilepsy started or
- have clinical features suggestive of a specific genetic epilepsy syndrome (for example, DS) or
- have additional clinical features such as:
- a learning disability
- autism spectrum disorder
- a structural abnormality (for example, dysmorphism or congenital malformation)
- unexplained cognitive or memory decline.
- Consider WGS for people with epilepsy of unknown cause who were aged between two and three years when epilepsy started, if clinically agreed by a specialist multidisciplinary team.50
United Kingdom Expert Group on Tuberous Sclerosis Complex
The expert group recommended, with consensus, to offer a genetic test at baseline to patients with definite or probable TSC. The group noted that a genetic test may clarify the diagnosis of TSC for patients that do not fulfill clinical criteria for the condition.51
In 2019, the group released recommendations for provision of coordinated care. They state, “with wider availability of genetic testing, identification of pathogenic mutations in TSC1 or TSC2 is now sufficient to establish a diagnosis, regardless of the presence of clinical features, and is particularly useful in confirming a suspected diagnosis, as many clinical TSC manifestations are infrequent in young patients.” The group further recommends that:
- For patients with newly diagnoses or suspected TSC:
- “Obtain three-generation family history to assess for additional family members at risk of TSC.
- Offer genetic testing for family counseling or when TSC diagnosis is in question but cannot be clinically confirmed.”
- For patients already diagnosed with definite or possible TSC:
- “Offer genetic testing for family counseling or when TSC diagnosis is in question but cannot be clinically confirmed.”52
Genetic Testing Advisory Committee (GTAC) for Ontario, Canada
These guidelines recommend the following as indications for genetic testing:
- “When the clinical features (age of onset, seizure semiology and EEG features) are consistent with a distinct epilepsy syndrome as defined by the International League Against Epilepsy (ILAE), with the exception of syndromes outlined in the following section.”
- “When the prognosis based on clinical and EEG findings is poor or the likelihood of lethal outcome is high.”
- “When epileptic seizures are refractory to medical treatment as defined by the ILAE12 (with no apparent acquired cause).”
- “When epilepsy is associated with features suggestive of inborn errors of metabolism.”
- “When epilepsy is associated with distinctive patterns of malformations of cortical development identified on neuroimaging studies.”
- “When epilepsy is associated with clinical signs of neurodegeneration.”
- “When epilepsy is associated with paroxysmal neurological features such as paroxysmal dyskinesias, episodic ataxias and hemiplegic migraine.”
- “When epilepsy is associated with additional syndromic features such as developmental delay/intellectual disability, multiple congenital anomalies and dysmorphic features.”
- “When familial epilepsy is present, defined as at least two first‐ degree family members with related epilepsy syndromes, unless the epilepsy syndrome is benign.”
The guidelines also recommend against genetic testing in the following situations:
- “Recognizable seizure syndrome with benign course.”
- “Childhood epilepsy with centro-temporal spikes (previously termed benign rolandic epilepsy).”
- “Isolated mesial temporal lobe epilepsy with hippocampal sclerosis.”
- “Typical childhood Absence epilepsy (although if it is early-onset or medically refractory epilepsy, one should consider and test for GLUT1 deficiency).”
- “Juvenile myoclonic epilepsy, which is well controlled on medications and without intellectual disability or any signs of neurodegeneration.”
- “Acquired epilepsy.”
The guideline also provides a list of “potentially treatable” genetic/metabolic epilepsies, shown below
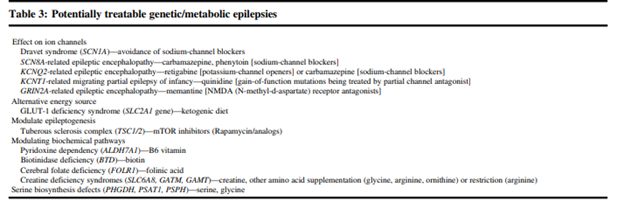
Finally, the guideline also makes recommendations for panel testing. Some situations in which a gene panel may be considered are if:
- epilepsy phenotyping is broad and does not easily classify the patient into a distinct group.
- “if the clinical diagnosis is clear and genetic heterogeneity is high, but clinical diagnosis is not indicative of a treatable condition.”
- “if clinical diagnosis is unclear and genetic heterogeneity is unknown.”
The guidelines further list examples of these situations, such as seizures with fever as a major trigger and idiopathic generalized epilepsy refractory to treatment.53
In addition to the indications for genetic testing outlined by the GTAC, the following recommendations are provided for the implementation of genetic panel testing in Ontario, Canada.
- It is essential that the genes selected have strong supporting evidence for their role in epilepsy. “It was recognized that genes present on commercially available panels did not, necessarily, show significant scientific evidence of being clinically relevant to epilepsy. Therefore, an evidence-based approach to the gene composition of a panel [...] was considered a priority”
- “The proposed epilepsy testing herein is based on a panel approach. This includes the option of a targeted or comprehensive panel approach or alternatively a filtered “exome” panel”
- Genetic panel testing may be considered in situations such as:
- “If the individual is young (<1 year) and does not present with an obvious recognizable syndrome, consider Panel 3 (Early Infantile Epileptic Encephalopathy Panel)
- If there is clinical suspicion of an 'actionable' gene, consider Panel 6 (STAT Option)
- If the individual is 1–3 years (approximately), nonsyndromic, with global developmental delay (GDD) or intellectual disability (ID), consider Panel 7 (Comprehensive Panel)”
- “An ongoing consultation process with local experts (clinical and laboratory), ad hoc working groups, and the end users of the panel test [...] will ensure performance outcomes of the epilepsy panels are captured and reviewed every 1–2 years”
- “For those with epilepsy plus additional comorbidities such as dysmorphic features, multisystem involvement, or progressive clinical course, an exome sequencing strategy may be the better initial test."54
References
- Berg AT, Berkovic SF, Brodie MJ, et al. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005-2009. Epilepsia. Apr 2010;51(4):676-85. doi:10.1111/j.1528-1167.2010.02522.x
- Myers CT, Mefford HC. Advancing epilepsy genetics in the genomic era. Genome medicine. Aug 25 2015;7:91. doi:10.1186/s13073-015-0214-7
- CDC. Epilepsy Facts and Stats. Updated May 15, 2024. https://www.cdc.gov/epilepsy/data-research/facts-stats/
- Williams CA, Battaglia A. Molecular biology of epilepsy genes. Experimental neurology. Jun 2013;244:51-8. doi:10.1016/j.expneurol.2011.12.001
- Ottman R, Hirose S, Jain S, et al. Genetic testing in the epilepsies--report of the ILAE Genetics Commission. Epilepsia. Apr 2010;51(4):655-70. doi:10.1111/j.1528-1167.2009.02429.x
- Hildebrand MS, Dahl HH, Damiano JA, Smith RJ, Scheffer IE, Berkovic SF. Recent advances in the molecular genetics of epilepsy. Journal of medical genetics. May 2013;50(5):271-9. doi:10.1136/jmedgenet-2012-101448
- Scheffer IE, Berkovic S, Capovilla G, et al. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017/04/01 2017;58(4):512-521. doi:10.1111/epi.13709
- Andrade D, Nascimento F. Dravet syndrome: Genetics, clinical features, and diagnosis. Updated June 19, 2024. https://www.uptodate.com/contents/dravet-syndrome-genetics-clinical-features-and-diagnosis
- Randle S. Tuberous sclerosis complex: Clinical features. Updated January 8, 2024. https://www.uptodate.com/contents/tuberous-sclerosis-complex-clinical-features
- Randle S. Tuberous sclerosis complex: Genetics and pathogenesis. Updated December 6, 2023. https://www.uptodate.com/contents/tuberous-sclerosis-complex-genetics-and-pathogenesis
- Randle S. Tuberous sclerosis complex: Evaluation and diagnosis. Updated February 29, 2024. https://www.uptodate.com/contents/tuberous-sclerosis-complex-evaluation-and-diagnosis
- Poduri A, Sheidley BR, Shostak S, Ottman R. Genetic testing in the epilepsies —developments and dilemmas. Nat Rev Neurol. May 2014;10(5):293-9. doi:10.1038/nrneurol.2014.60
- Heron SE, Regan BM, Harris RV, et al. Association of SLC32A1 Missense Variants With Genetic Epilepsy With Febrile Seizures Plus. Neurology. May 4 2021;96(18):e2251-e2260. doi:10.1212/wnl.0000000000011855
- Lemke JR, Riesch E, Scheurenbrand T, et al. Targeted next generation sequencing as a diagnostic tool in epileptic disorders. Epilepsia. Aug 2012;53(8):1387-98. doi:10.1111/j.1528-1167.2012.03516.x
- Moller RS, Larsen LH, Johannesen KM, et al. Gene Panel Testing in Epileptic Encephalopathies and Familial Epilepsies. Molecular syndromology. Sep 2016;7(4):210-219. doi:10.1159/000448369
- Stenhouse SA, Ellis R, Zuberi S. SCN1A Genetic Test for Dravet Syndrome (Severe Myoclonic Epilepsy of Infancy and its Clinical Subtypes) for use in the Diagnosis, Prognosis, Treatment and Management of Dravet Syndrome. PLoS currents. 2013;5doi:10.1371/currents.eogt.c553b83d745dd79bfb61eaf35e522b0b
- Mefford HC. Clinical Genetic Testing in Epilepsy. Epilepsy currents. Jul-Aug 2015;15(4):197-201. doi:10.5698/1535-7511-15.4.197
- Hrabik SA, Standridge SM, Greiner HM, et al. The Clinical Utility of a Single-Nucleotide Polymorphism Microarray in Patients With Epilepsy at a Tertiary Medical Center. Journal of child neurology. Nov 2015;30(13):1770-7. doi:10.1177/0883073815579972
- Olson H, Shen Y, Avallone J, et al. Copy number variation plays an important role in clinical epilepsy. Annals of neurology. Jun 2014;75(6):943-58. doi:10.1002/ana.24178
- Berg AT, Coryell J, Saneto RP, et al. Early-Life Epilepsies and the Emerging Role of Genetic Testing. JAMA pediatrics. Sep 1 2017;171(9):863-871. doi:10.1001/jamapediatrics.2017.1743
- Dunn P, Albury CL, Maksemous N, et al. Next Generation Sequencing Methods for Diagnosis of Epilepsy Syndromes. Frontiers in genetics. 2018;9:20. doi:10.3389/fgene.2018.00020
- Stosser MB, Lindy AS, Butler E, et al. High frequency of mosaic pathogenic variants in genes causing epilepsy-related neurodevelopmental disorders. Genetics in medicine : official journal of the American College of Medical Genetics. Aug 24 2017;doi:10.1038/gim.2017.114
- Poduri A. When Should Genetic Testing Be Performed in Epilepsy Patients? Epilepsy currents. Jan-Feb 2017;17(1):16-22. doi:10.5698/1535-7511-17.1.16
- Falsaperla R, Corsello G. Pyridoxine dependent epilepsies: new therapeutical point of view. Italian journal of pediatrics. 2017;43(1):68. doi:10.1186/s13052-017-0387-3
- Peng J, Zhou Y, Wang K. Multiplex gene and phenotype network to characterize shared genetic pathways of epilepsy and autism. Sci Rep. Jan 13 2021;11(1):952. doi:10.1038/s41598-020-78654-y
- Long S, Zhou H, Li S, et al. The Clinical and Genetic Features of Co-occurring Epilepsy and Autism Spectrum Disorder in Chinese Children. Front Neurol. 2019;10:505. doi:10.3389/fneur.2019.00505
- Sanchez Fernandez I, Loddenkemper T, Gainza-Lein M, Sheidley BR, Poduri A. Diagnostic yield of genetic tests in epilepsy: A meta-analysis and cost-effectiveness study. Neurology. Jan 4 2019;doi:10.1212/wnl.0000000000006850
- Borlot F, de Almeida BI, Combe SL, Andrade DM, Filloux FM, Myers KA. Clinical utility of multigene panel testing in adults with epilepsy and intellectual disability. Epilepsia. Aug 2019;60(8):1661-1669. doi:10.1111/epi.16273
- Kim SY, Jang SS, Kim H, et al. Genetic diagnosis of infantile-onset epilepsy in the clinic: Application of whole-exome sequencing following epilepsy gene panel testing. Clinical Genetics. 2021;99(3):418-424. doi:10.1111/cge.13903
- Willimsky EK, Munzig A, Mayer K, et al. Next Generation Sequencing in Pediatric Epilepsy Using Customized Panels: Size Matters. Neuropediatrics. Apr 2021;52(2):92-97. doi:10.1055/s-0040-1712488
- Stefanski A, Calle-López Y, Leu C, Pérez-Palma E, Pestana-Knight E, Lal D. Clinical sequencing yield in epilepsy, autism spectrum disorder, and intellectual disability: A systematic review and meta-analysis. Epilepsia. Jan 2021;62(1):143-151. doi:10.1111/epi.16755
- Sheidley BR, Malinowski J, Bergner AL, et al. Genetic testing for the epilepsies: A systematic review. Epilepsia. Dec 10 2021;doi:10.1111/epi.17141
- Barbosa M, Joshi RS, Garg P, et al. Identification of rare de novo epigenetic variations in congenital disorders. Nat Commun. May 25 2018;9(1):2064. doi:10.1038/s41467-018-04540-x
- Hebbar M, Mefford HC. Recent advances in epilepsy genomics and genetic testing. F1000Res. 2020;9doi:10.12688/f1000research.21366.1
- Balestrini S, Sisodiya SM. Pharmacogenomics in epilepsy. Neurosci Lett. Feb 22 2018;667:27-39. doi:10.1016/j.neulet.2017.01.014
- McKnight D, Bristow SL, Truty RM, et al. Multigene panel testing in a large cohort of adults with epilepsy: diagnostic yield and clinically actionable genetic findings. Neurology Genetics. 2022;8(1)doi:10.1212/NXG.0000000000000650
- Wilmshurst JM, Gaillard WD, Vinayan KP, et al. Summary of recommendations for the management of infantile seizures: Task Force Report for the ILAE Commission of Pediatrics. Epilepsia. Aug 2015;56(8):1185-97. doi:10.1111/epi.13057
- Norman M, Wilson SJ, Baxendale S, et al. Addressing neuropsychological diagnostics in adults with epilepsy: Introducing the International Classification of Cognitive Disorders in Epilepsy: The IC CODE Initiative. Epilepsia Open. Jun 2021;6(2):266-275. doi:10.1002/epi4.12478
- Zuberi SM, Wirrell E, Yozawitz E, et al. ILAE classification and definition of epilepsy syndromes with onset in neonates and infants: Position statement by the ILAE Task Force on Nosology and Definitions. Epilepsia. Jun 2022;63(6):1349-1397. doi:10.1111/epi.17239
- Hirsch E, French J, Scheffer IE, et al. ILAE definition of the Idiopathic Generalized Epilepsy Syndromes: Position statement by the ILAE Task Force on Nosology and Definitions. Epilepsia. 2022;63(6):1475-1499. doi:10.1111/epi.17236
- Burgunder JM, Finsterer J, Szolnoki Z, et al. EFNS guidelines on the molecular diagnosis of channelopathies, epilepsies, migraine, stroke, and dementias. European Journal of Neurology. 2010/05/01 2010;17(5):641-648. doi:10.1111/j.1468-1331.2010.02985.x
- Ferrazzoli C. Genetic testing in adults with epilepsy. Scientific Panel Epilepsy. Updated June 4, 2024. https://www.ean.org/research/resources/neurology-updates/detail/genetic-testing-in-adults-with-epilepsy
- Riviello JJ, Ashwal S, Hirtz D, et al. Practice Parameter: Diagnostic assessment of the child with status epilepticus (an evidence-based review). Neurology. 2006;67(9):1542. doi:10.1212/01.wnl.0000243197.05519.3d
- More than Four in 10 Adults with Epilepsy Have a Genetic Link, Adults with Unknown Cause of Condition Should Get Tested, Study Suggests. December 1, 2023, https://aesnet.org/about/aes-press-room/press-releases/more-than-four-in-10-adults-with-epilepsy-have-a-genetic-link-adults-with-unknown-cause-of-condition-should-get-tested-study-suggests
- Smith L, Malinowski J, Ceulemans S, et al. Genetic testing and counseling for the unexplained epilepsies: An evidence-based practice guideline of the National Society of Genetic Counselors. Journal of Genetic Counseling. 2023;32(2):266-280. doi:10.1002/jgc4.1646
- Klepper J, Akman C, Armeno M, et al. Glut1 Deficiency Syndrome (Glut1DS): State of the art in 2020 and recommendations of the international Glut1DS study group. Epilepsia open. 2020;5(3):354-365. doi:10.1002/epi4.12414
- Wirrell EC, Laux L, Donner E, et al. Optimizing the Diagnosis and Management of Dravet Syndrome: Recommendations From a North American Consensus Panel. Pediatric neurology. Mar 2017;68:18-34.e3. doi:10.1016/j.pediatrneurol.2017.01.025
- Dravet Syndrome Foundation. The Diagnosis and Treatment of Dravet Syndrome. https://dravetfoundation.org/hcp-resources/diagnosis-and-treatment/
- SIGN. Diagnosis and management of epilepsy in adults. https://www.sign.ac.uk/media/1079/sign143_2018.pdf
- NICE. Epilepsies in children, young people and adults. Updated January 30. https://www.nice.org.uk/guidance/ng217
- Amin S, Kingswood JC, Bolton PF, et al. The UK guidelines for management and surveillance of Tuberous Sclerosis Complex. QJM: An International Journal of Medicine. 2018;112(3):171-182. doi:10.1093/qjmed/hcy215
- Annear NMP, Appleton RE, Bassi Z, et al. Tuberous Sclerosis Complex (TSC): Expert Recommendations for Provision of Coordinated Care. Front Neurol. 2019;10:1116. doi:10.3389/fneur.2019.01116
- Jain P, Andrade D, Donner E, et al. Development of Criteria for Epilepsy Genetic Testing in Ontario, Canada. The Canadian journal of neurological sciences Le journal canadien des sciences neurologiques. Jan 2019;46(1):7-13. doi:10.1017/cjn.2018.341
- Dyment DA, Prasad AN, Boycott KM, et al. Implementation of Epilepsy Multigene Panel Testing in Ontario, Canada. Canadian Journal of Neurological Sciences / Journal Canadien des Sciences Neurologiques. 2020;47(1):61-68. doi:10.1017/cjn.2019.304
Coding Section
| Codes | Number | Description |
| 81404 | Molecular pathology procedure, Level 5 (eg, analysis of 2-5 exons by DNA sequence analysis, mutation scanning or duplication/deletion variants of 6-10 exons, or characterization of a dynamic mutation disorder/triplet repeat by Southern blot analysis) | |
| 81405 | Molecular pathology procedure, Level 6 (eg, analysis of 6-10 exons by DNA sequence analysis, mutation scanning or duplication/deletion variants of 11-25 exons, regionally targeted cytogenomic array analysis) | |
| 81406 | Molecular pathology procedure, Level 7 (eg, analysis of 11-25 exons by DNA sequence analysis, mutation scanning or duplication/deletion variants of 26-50 exons) | |
| 81407 | Molecular pathology procedure, Level 8 (eg, analysis of 26-50 exons by DNA sequence analysis, mutation scanning or duplication/deletion variants of >50 exons, sequence analysis of multiple genes on one platform) | |
| 81415 | Exome (eg, unexplained constitutional or heritable disorder or syndrome); sequence analysis | |
| 81416 | Exome (eg, unexplained constitutional or heritable disorder or syndrome); sequence analysis, each comparator exome (eg, parents, siblings) (List separately in addition to code for primary procedure) | |
| 81419 (effective 01/01/2020) | Epilepsy genomic sequence analysis panel, must include analyses for ALDH7A1, CACNA1A, CDKL5, CHD2, GABRG2, GRIN2A, KCNQ2, MECP2, PCDH19, POLG, PRRT2, SCN1A, SCN1B, SCN2A, SCN8A, SLC2A1, SLC9A6, STXBP1, SYNGAP1, TCF4, TPP1, TSC1, TSC2, and ZEB2 | |
| 81425 | Genome (eg, unexplained constitutional or heritable disorder or syndrome); sequence analysis | |
| 81426 | Genome (eg, unexplained constitutional or heritable disorder or syndrome); sequence analysis, each comparator genome (eg, parents, siblings) (List separately in addition to code for primary procedure) | |
| 81479 | Unlisted molecular pathology procedure | |
| ICD-9-CM Diagnosis | ||
| 345.00-345.91 | epilepsy code range | |
| ICD-10-CM (effective 10/01/15) | ||
| G40.001-G40.919 | Epilepsy code range | |
| ICD-10-PCS (effective 10/01/15) | Not applicable. ICD-10-PCS codes are only used for inpatient services. There are no ICD procedure codes for laboratory test. |
Procedure and diagnosis codes on Medical Policy documents are included only as a general reference tool for each policy. They may not be all-inclusive.
Index
Epilepsy, Genetic Testing
This medical policy was developed through consideration of peer-reviewed medical literature generally recognized by the relevant medical community, U.S. FDA approval status, nationally accepted standards of medical practice and accepted standards of medical practice in this community and other nonaffiliated technology evaluation centers, reference to federal regulations, other plan medical policies and accredited national guidelines.
"Current Procedural Terminology © American Medical Association. All Rights Reserved"
History From 2013 Forward
| 04/15/2025 | Annual review, adding criteria #1 requiring genetic counseling, updating criteria #2 to remove age restriction and allows for WES or multi-gene panel testing. Adding new WES criteria. Updating note 1. Also updating description, table of terminology, rationale, guidelines/recommendations and references. Adding codes 81415, 81416, 81425, 81426. |
| 08/01/2024 | Annual review, no change to policy intent. Updating rationale, references. Note 1 directs reader to CAM 235 updating verbiage for CPT 81406. |
| 04/25/2024 | Moving review date to 7/2024. Review will coinside with Avalon's review date. |
| 04/03/2023 | Annual review no change to policy intent, but policy is being rewritten for clarity and consistency. Also updating rational, references and table of terminology. |
| 04/08/2022 |
Annual review, no change to policy intent. Updating rationale and references. Adding table of terminology. |
| 04/01/2021 |
Annual review, no change to policy intent. Updating background, rationale and references. |
| 12/10/2020 |
Updating Coding Section with 2021 codes |
| 04/16/2020 |
Annual review, updating policy to include multiple additional genes for testing. No other changes. |
| 04/04/2019 |
Annual review, no change to policy intent. |
| 04/17/2018 |
Annual review, no change to policy intent. |
| 04/17/2017 |
Annual review, updating policy to allow for PCDH19 testing. Updated category to Laboratory. No other changes |
| 01/05/2017 |
Annual review, no change to intent of policy. |
| 01/21/2016 |
Annual review, no change to policy intent. Updating background, description, regulatory status, related policies, guidelines, rationale, references and coding. |
| 01/04/2016 |
Updated CPT coding |
| 01/20/2015 |
Annual review, no change to policy intent. Added coding. |
| 01/07/2014 |
NEW POLICY |