Molecular Diagnostics for Breast Cancer Prognosis - CAM 278
Description:
Molecular diagnostics for breast cancer management is emerging as a useful tool to aid in disease prognosis or therapeutic efficacy on an individual level. Many of the currently available molecular tests are gene expression assays that measure the number of transcribed copies of specific mRNAs to assess the genes that are active in tumor tissue, which can be clinically useful for disease classification, diagnosis, and prognosis, as well as for tailoring treatment to underlying genetic determinants of pharmacologic response.1
Policy
Application of coverage criteria is dependent upon an individual’s benefit coverage at the time of the request.
- For individuals with newly diagnosed breast cancer, tumor testing for hormone receptor (estrogen receptor and progesterone receptor) expression and human epidermal growth factor receptor 2 (HER2) overexpression is considered MEDCIALLY NECESSARY.
- For individuals who have been diagnosed with invasive breast cancer and who are being considered for adjuvant chemotherapy, use of the Oncotype DX 21-gene expression assay (see Note 1) is considered MEDCIALLY NECESSARY when all of the following criteria are also met:
- Histology that is ductal/no special type (NST) (see Note 2), lobular, mixed or micropapillary.
- pT1, pT2, or pT3 cancer.
- Hormone receptor (HR) positive (either receptor [ER] or progesterone-receptor [PR] positive) cancer.
- Human epidermal growth factor receptor two (HER2) negative cancer.
- Tumor is one of the following:
- pN0 tumor that is > 0.5 cm in size.
- pN1mi (≤ 2 mm axillary node metastases).
- pN1 (1 – 3 positive nodes).
- For individuals who have been diagnosed with pT1, pT2, or operable pT3, HR positive, HER2-negative, and node-negative or pN1 (1 – 3 positive nodes) breast cancer and who have been determined to be at high clinical risk per MINDACT categorization (see Note 3), use of the MammaPrint assay is considered MEDCIALLY NECESSARY.
- For individuals who have been diagnosed with T1 or T2, HR positive, HER2-negative, and node-negative or pN1 (1 – 3 positive nodes) breast cancer, use of the 12-gene (EndoPredict) assay or the 50-gene (Prosigna) assay is considered MEDCIALLY NECESSARY.
- For individuals who have been diagnosed with T1-T3, HR-positive, HER2-negative breast cancer, use of the Breast Cancer Index (BCI) test to determine the benefit of extended adjuvant endocrine therapy is considered MEDCIALLY NECESSARY.
The following does not meet coverage criteria due to a lack of available published scientific literature confirming that the test(s) is/are required and beneficial for the diagnosis and treatment of an individual’s illness.
- In males, use of gene expression assays other than 21-gene RT-PCR-based assays is considered NOT MEDCIALLY NECESSARY.
- Use of gene expression assays not mentioned above (e.g., Mammostrat, BluePrint®) is considered NOT MEDCIALLY NECESSARY.
- The use of Oncotype DX for ductal carcinoma in situ (DCIS) is considered NOT MEDCIALLY NECESSARY.
- For all other indications or in any other situation not mentioned above, use of gene expression assays is considered NOT MEDCIALLY NECESSARY.
- Use of protein profiling assays (e.g., DCISionRT®, BBDRisk Dx®) is considered NOT MEDCIALLY NECESSARY
NOTES:
Note 1: The assay should only be ordered on a tissue specimen obtained during surgical removal of the tumor and after subsequent pathology examination of the tumor has been completed and determined to meet the above criteria (i.e., the test should not be ordered on a preliminary core biopsy without surgical staging). For patients who have multiple primary tumors, a specimen from the tumor with the most aggressive histological characteristics should be submitted for testing. It is not necessary to conduct testing on each tumor; treatment is based on the most aggressive lesion.
Note 2: As per NCCN Clinical Practice Guidelines in Oncology (Breast Cancer), “According to WHO, carcinoma of no special type (NST) encompasses multiple patterns including medullary pattern, cancers with neuroendocrine expression, and other rare patterns” (NCCN, 2024).
Note 3: As per NCCN Clinical Practice Guidelines in Oncology (Breast Cancer), “High clinical risk in the MINDACT trial was defined for grade 1 tumors as > 3 cm N0 or T2N1, for grade 2 tumors T2N0-1, and for grade 3 tumors T1c-2N0-1” (NCCN, 2024).
Table of Terminology
| Term |
Definition |
| ABHD14A |
Abhydrolase domain containing 14A gene |
| ACS |
American Cancer Society |
| AI |
Aromatase inhibitor |
| ASCO |
American Society of Clinical Oncology |
| ATP6V0A4 |
ATPase H+ transporting V0 subunit a4 gene |
| AUC |
Areas under the curve |
| AZGP1 |
Alpha-2-glycoprotein 1, zinc-binding gene |
| BCI |
Breast cancer index |
| BEND3 |
BEN domain containing 3 gene |
| BIRC5 |
Baculoviral IAP repeat containing 5 gene |
| BTG2 |
BTG anti-proliferation factor 2 gene |
| BUB1B |
BUB1 mitotic checkpoint serine/threonine kinase B gene |
| CDYL |
Chromodomain Y like gene |
| CEF |
Cyclophosphamide, epirubicin and fluorouracil |
| CENPA |
Centromere protein A |
| CET |
Chemoendocrine Therapy |
| CHAD |
Chondroadherin gene |
| CHER-LOB |
Chemotherapy, erceptin, and lapatinib in operable breast cancer |
| CMF |
Cyclophosphamide, methotrexate and fluorouracil |
| CNB |
Core needle biopsy |
| CTCs |
Circulating tumour cells |
| CTS |
Clinical treatment score |
| DCIS |
Ductal carcinoma in situ |
| DHCR7 |
7-dehydrocholesterol reductase gene |
| DRFR |
Distant recurrence free rate |
| EBC |
Early HER2-negative breast cancer |
| EET |
Extended Endocrine Therapy |
| EGAPP |
Evaluation of Genomic Applications in Practice and Prevention |
| EGTM |
European Group on Tumour Markers |
| ER |
Estrogen receptor |
| ESMO |
European Society for Medical Oncology |
| ET |
Endocrine therapy |
| FDA |
Food and Drug Association |
| FNAB |
Fine-needle aspirate biopsy |
| HER2 |
Human epidermal growth factor receptor two |
| HER2DX |
Model for predicting disease-free survival |
| HR |
Hazard ratio |
| IDFS |
Invasive disease-free survival |
| IDUA |
Alpha-L-iduronidase gene |
| IHC4 |
Immunohistochemistry 4 |
| IL6ST |
Interleukin 6 cytokine family signal transducer gene |
| InvBE |
Ipsilateral breast event |
| KIF18A |
Kinesin family member 18A gene |
| LN |
Lymph node |
| MFS |
Metastasis free survival |
| MGI |
Molecular grade index |
| MGP |
Matrix Gla protein gene |
| MINDACT |
Microarray in node negative disease may avoid chemotherapy |
| mRNAs |
Messenger ribonucleic acid |
| MYL5 |
Myosin light chain 5 gene |
| NaCT |
Neoadjuvant chemotherapy |
| NCCN |
National Comprehensive Cancer Network |
| NEK2 |
NIMA related kinase 2 gene |
| NET |
Neoadjuvant endocrine therapy |
| NICE |
National Institute for Health and Care Excellence |
| NPI |
Nottingham prognostic index |
| NST |
No special type |
| PAI-1 |
Plasminogen Activator Inhibitor Type 1 |
| PAM50 |
Prosigna assay looking at the activity of 50 genes |
| PMBC |
Patients with pure mucinous breast cancer |
| PR |
Progesterone Receptor |
| PREP |
Prolyl endopeptidase |
| RACGAP1 |
Rac GTPase activating protein 1 gene |
| RBBP8 |
RB binding protein 8, endonuclease gene |
| RCB |
Residual cancer burden |
| ROR |
Risk of recurrence |
| RRM2 |
Ribonucleotide reductase regulatory subunit M2 gene |
| RS |
Recurrence Score |
| RTN4IP1 |
Reticulon 4 interacting protein 1 gene |
| RT-PCR |
Reverse transcriptase/transcription polymerase chain reaction |
| S100PBP |
S100P binding protein gene |
| STC2 |
Stanniocalcin 2 gene |
| TAILORx |
Trial assigning individualized options for treatment (Rx) |
| TotBE |
Total breast event |
| TPRG1 |
Tumor protein p63 regulated 1 gene |
| UBE2C |
Ubiquitin conjugating enzyme E2 C gene |
| uPA |
Urokinase plasminogen activator |
| WDR6 |
WD repeat domain 6 gene |
Rationale
Breast cancer is the most frequently diagnosed cancer and is the leading cause of cancer death globally in women. Approximately one in eight women will develop breast cancer in their lifetime.3,4
While adjuvant systemic therapy has reduced mortality from breast cancer,5-8 it is not without its risks and costs; therefore, reliable prognostic profiles for recurrence and clinically applicable predictive factors are of great value in the management of the disease.9
Several biology-based prognostic profiles have been developed, validated, and are in clinical use to predict breast cancer recurrence or response to chemotherapy. Among the most useful biomarkers for this purpose are the estrogen receptors (ER), progesterone receptors (PR), and the human epidermal growth factor receptor two (HER2). Tumors that express one or more of these proteins may have different sensitivities to certain cytotoxic chemotherapeutic agents, influencing the strategic management of the cancer. Moreover, many proprietary panels that aim to provide predictive and prognostic information about an individual’s cancer are only suitable for use in tumors of a particular type (i.e., HER2-negative). Intensive research efforts are ongoing to refine the clinical utility and the indications for these predictive and prognostic profiles.10 In addition, as next generation sequencing of tumor genomes progresses, these profiles will be improved or replaced by the next generation of molecular profiles.9
Chen, et al. (2018) examined the association of genomic methylation and the changes in the subsequent transcriptome. The authors observed how a chronic condition influenced epigenomic changes. A human volunteer provided peripheral blood samples over 36 months and the authors created 28 methylome datasets as well as 57 transcriptome datasets. During this period, the human volunteer experienced six viral infections and two elevated periods of fasting glucose and glycated hemoglobin A1c. The authors noted that the methylome changes correlated with the glucose level changes and that the gene expression levels varied greatly, often during the viral infections. The authors proposed that the DNA methylation was the primary gene expression mechanism for chronic conditions, as it was generally a stable epigenetic marker.11
Rueda, et al. (2019) presented a disease model that stratifies distinct stages and incorporated factors such as “locoregional recurrence, distant recurrence, breast-cancer-related death and death from other causes.”12 This model integrated these features into an individual risk of recurrence prediction and was applied to 3240 patients, of which 1980 had molecular data. From this model, the authors identified four late-recurring “integrative subtypes, comprising about one quarter (26%) of tumours that are both positive for ER and negative for human epidermal growth factor receptor 2, each with characteristic tumour-driving alterations in genomic copy number and a high risk of recurrence (mean 47–62%) up to 20 years after diagnosis.” These four subtypes were enriched in copy number alterations, and these alterations were labeled the likely drivers of each subgroup. A triple negative subgroup, IntCluster 10, was found to be largely relapse-free after five years whereas the IntCluster4 ER negative subgroup was found to have a large risk of recurrence.12
Innovative ideas continue to spread as the field burgeons, not limiting its inspiration to etiology. Savci-Heijink, et al. (2019) believe in the clinical value of identifying a gene expression profile in tumors that selects a subpopulation of patients that is more likely to develop visceral organ metastases due to their association with poor survival outcomes in breast cancers. The researchers analyzed the genetic profiles of 157 primary tumors from breast cancer patients who developed distant metastases and differentially expressed genes between the group of tumors with visceral metastasis and those without visceral metastases were noted. The results yielded fourteen differentially expressed genes (WDR6, CDYL, ATP6V0A4, CHAD, IDUA, MYL5, PREP, RTN4IP1, BTG2, TPRG1, ABHD14A, KIF18A, S100PBP and BEND3) between the tumors with visceral metastasis and those without visceral metastases. According to the data, five of the genes (CDYL, ATP6V0A4, PREP, RTN4IP1 and KIF18A) were upregulated while the other genes were downregulated, creating a list of biomarkers and genetic signatures of interest that may result in earlier diagnosis and treatment.13
Proprietary Testing
OncoType DX
The Oncotype Dx 21-gene recurrence score (RS) is the best validated prognostic assay and may identify patients who are most and least likely to derive benefit from adjuvant chemotherapy. The expression levels of 16 genes (plus five reference genes) are measured by quantitative reverse transcription polymerase chain reaction (RT-PCR). The sum of this calculation is known as the RS to optimize prediction of distant relapse despite tamoxifen therapy. At this time, it is indicated for individuals with node-negative, estrogen receptor (ER)-positive, human epidermal growth factor receptor 2 (HER2)-negative breast cancer to determine the prognosis in patients recommended to proceed with at least a five-year course of endocrine therapy. The optimal RS cutoff for omission of chemotherapy remains unclear given that the different studies have used different cutoffs.14-17 However, it may be reasonable not to administer adjuvant chemotherapy for patients with node-negative, ER-positive breast cancer and an RS of <15.9
Sparano, et al. (2018) performed a prospective trial to assess the utility of the recurrence score based on the 21 gene breast cancer assay to predict chemotherapy in patients who have a midrange score. “Of the 9719 eligible patients with follow-up information, 6711 (69%) had a midrange recurrence score of 11 to 25 and were randomly assigned to receive either chemoendocrine therapy or endocrine therapy alone. The trial was designed to show noninferiority of endocrine therapy alone for invasive disease-free survival (defined as freedom from invasive disease recurrence, second primary cancer, or death).” They found that “Endocrine therapy was noninferior to chemoendocrine therapy in the analysis of invasive disease-free survival (hazard ratio for invasive disease recurrence, second primary cancer, or death [endocrine vs. chemoendocrine therapy], 1.08; 95% confidence interval, 0.94 to 1.24; P=0.26). At nine years, the two treatment groups had similar rates of invasive disease-free survival (83.3% in the endocrine-therapy group and 84.3% in the chemoendocrine-therapy group), freedom from disease recurrence at a distant site (94.5% and 95.0%) or at a distant or local-regional site (92.2% and 92.9%), and overall survival (93.9% and 93.8%). The chemotherapy benefit for invasive disease-free survival varied with the combination of recurrence score and age (P=0.004), with some benefit of chemotherapy found in women 50 years of age or younger with a recurrence score of 16 to 25.” They concluded that “Adjuvant endocrine therapy and chemoendocrine therapy had similar efficacy in women with hormone-receptor-positive, HER2-negative, axillary node-negative breast cancer who had a midrange 21-gene recurrence score, although some benefit of chemotherapy was found in some women 50 years of age or younger.”18
In a prospective, randomized trial of endocrine therapy (ET) versus chemoendocrine therapy (CET), Kalinsky, et al. (2021) attempted to gather support for the potential prognostic and predictive role of the RS for CT benefit in postmenopausal patients with one to three lymph node-positive breast cancer using Oncotype Dx in the RxPonder Trial. In the study, women with a RS less than 25 were randomized to receive ET or CET in 1:1 randomization using 3 stratification factors—RS (0-13 vs.14-25), menopausal status, and axillary nodal dissection vs. sentinel node biopsy—with the end goal of ascertaining whether the effect of CT on invasive disease-free survival (IDFS) was a function of RS. Of 9,383 patients screened in the study, 5,083 patients were randomized based on having met eligibility criteria and 5,018 participated in the trial. With a median follow-up period of 5.1 years, the authors observed 447 IDFS events. It was reported that in a model with CT, RS, and menopausal status (no interaction term), higher continuous RS was associated with worse IDFS (hazard ratio HR 1.06, 2-sided p<0) and that and CT was associated with an improvement in IDFS (HR 0.81, p=0.026). Moreover, a distinction exists between postmenopausal and premenopausal patients in their reception of chemoendocrine versus endocrine therapy based on their RS: in postmenopausal patients (n=3350), the HR for CET vs. ET was not significant (HR=0.97, p=0.82, indicating no benefit from CT, whereas in premenopausal patients (N=1665, 33%), the HR (0.54) was statistically significant (p=0.0004), suggesting CT benefit. In other words, postmenopausal patients in this situation may be spared adjuvant chemotherapy, while premenopausal patients have much to benefit from the same. Therefore, the authors concluded that “the current data show that adjuvant therapy can be de-escalated to ET alone in postmenopausal pts with a RS < 25 and 1-3 +LN.”19
Lashen, et al. (2023) studied the performance of Oncotype DX compared to conventional clinicopathological parameters. The study included 420 estrogen receptor-positive/HER2-negative breast cancer patients in the United Kingdom. All patients included in the study had undergone Oncotype DX testing. The authors found a significant association between a high Oncotype DX recurrence score and grade 3 tumors, as well as a significant association between low Oncotype DX recurrence score and special tumor types. Additionally, there was an inverse association between Oncotype DX recurrence score and the levels of estrogen receptor and progesterone receptor expression, and a positive linear relationship between Oncotype DX recurrence score and Ki67 labelling index. The authors state that “some tumour features such as tumour grade, type, PR status and Ki67 index can be used as surrogate markers in certain scenarios.” The authors concluded that “Oncotype DX RS is correlated strongly with the conventional clinicopathological parameters.”20
EndoPredict
EndoPredict® (Myriad Genetic, Inc.) is a breast cancer prognostic test that assesses the expression of eight target genes as compared to three housekeeping (normalization) genes and one additional control gene to detect contamination by residual DNA. The results of this testing are combined to produce a clinical score to determine whether the breast cancer is at high- or low-risk of possible recurrence within ten years.21,22
According to the Myriad EndoPredict® Technical Specifications (Effective Date: December 21, 2021), if the test is used on resected tissue, the 12-gene molecular score is then “combined with tumor size and lymph node status to generate an EPclin Risk Score associated recurrence risks and estimated absolute benefit of chemotherapy” whereas “the molecular score and its associated 10-year risk of distance recurrence…are provided” if the sample is a biopsy specimen.23 The target genes in the EndoPredict® test include the following:22,24
- BIRC5
- DHCR7
- UBE2C
- AZGP1
- IL6ST
- MGP
- RBBP8
- STC2
Filipits, et al. (2011) validated EP’s 11 gene version (only missing the one gene to control for DNA contamination, HBB). The investigators combined these gene expression results with nodal status and tumor size into a risk score (“EPclin”). Two cohorts of 378 and 1324 samples were used in this study, and both cohorts demonstrated that the risk score produced was an independent predictor of distant recurrence. The investigators calculated that EPclin had superior prognostic power compared to solely clinicopathologic factors. EPClin was also found to result in a 4% rate of recurrence for both low-risk cohorts in each sample, a 28% recurrence rate in the “high risk” portion of the 378-item sample, and a 22% recurrence rate in the “high risk” portion of the 1324-item sample. The authors concluded that “the multigene EP risk score provided additional prognostic information to the risk of distant recurrence of breast cancer patients, independent from clinicopathologic parameters. The EPclin score outperformed all conventional clinicopathologic risk factors.25,26
Two studies by Sestak, et al. (2018)have also shown the clinical validity and utility of EndoPredict® testing in women with node-positive breast cancer.27,28 One study consisted of both node-positive and node-negative women (n=3746) to compare the performance of EPclin for predicting distant recurrence in women who had either received endocrine therapy (ET) alone or ET with chemotherapy (ET+C). “In this comparative non-randomised analysis, the rate of increase in DR with EPclin score was significantly reduced in women who received ET + C versus ET alone. Our indirect comparisons suggest that a high EPclin score can predict chemotherapy benefit in women with ER-positive, HER2-negative disease.”28 In the other study, the researchers compared the performance of six different prognostic signatures for ER-positive breast cancer in two different patient groups—node-negative and node-positive. The six tests include EndoPredict®, Breast Cancer Index (BCI), Clinical Treatment Score (CTS), IHC4, ROR, and RS. Within the node-positive cohort (n=277), the authors report the highest univariate hazard ratio (HR) for EPclin (1.69) compared to 1.39 for RS.27
Another study (n=1702) compared the use of EndoPredict® in node-positive and node-negative women treated with endocrine therapy only. This study assessed both the 10- and 15-year distant recurrence-free rate (DRFR). Similar to the node-negative individuals, the 10-year DRFR for patients with one to three positive nodes improved significantly higher than for the full cohort (95.6% versus 80.9%, respectively). “The molecular and EPclin scores were significant predictors of DRFR after adjusting for clinical variables, regardless of nodal status.”24
Evidence of EndoPredict’s ability to produce a change in therapeutic management abound. For example, Villarreal-Garza, et al. (2020) enrolled 99 premenopausal women with a median age of 43 and with hormone receptor-positive, HER2-negative, T1-T2, and N0-N1 breast cancer, and the cases were presented to a multidisciplinary tumor board. The researchers reported that a “change in chemotherapy or endocrine therapy regimen was suggested in 19% and 20% of cases” when EPclin results were made available. Moreover, it was reported that a significant difference could be “found in the median EPclin score between patients with a low- vs. high-intensity chemotherapy and endocrine therapy regimen recommendation (p = 0.049 and p = 0.0001, respectively)”, which may have contributed to high final treatment adherence (93%) in premenopausal patients.29
Dubsky, et al. (2020) used hormone receptor (HR)-positive, HER2-negative samples from patients in the ABCSG-34 randomized phase II trial to generate a 12-gene molecular score with the goal of assessing EndoPredict’s ability to predict response to neoadjuvant chemotherapy (NaCT) or neoadjuvant endocrine therapy (NET). The patients in the study were assigned to receive either NaCT or NET based on menopausal status, HR expression, grade and Ki67, and their responses to their assigned therapies were measured by using residual cancer burden (RCB). It was reported that the 12-gene molecular score significantly predicted treatment response for NaCT and NET, boasting areas under the curve of 0.736 and 0.726 respectively. Based on these results, it was concluded that “Tumours with low MS were unlikely to benefit from NaCT, whereas a high MS predicted resistance to NET”, further motivating the use of EndoPredict to provide personalized treatment to patients.30
Breast Cancer Index
The Breast Cancer Index (BCI) is a combination of two profiles, the HOXB13-to-IL17BR expression ratio (H:I ratio) and the Molecular Grade Index (MGI). This combination is intended to report “Whether or not an additional five years of anti-estrogen therapy is likely to help reduce [the] risk of the cancer returning” and “[the] individual risk of recurrence after 5 years based on [the] original tumor biology.” The five genes the MGI examines are BUB1B, CENPA, NEK2, RACGAP1, and RRM2.31 Using genome-wide microarray analysis, three differentially expressed genes that were associated with an increased risk of progression among ER-positive patients treated with tamoxifen were: the antiapoptotic homeobox B13 (HOXB13, overexpressed in tamoxifen recurrent cases) and both interleukin 17B receptor (IL17BR) and EST AI240933 (both overexpressed in tamoxifen nonrecurrent cases).9
The MGI was validated by separate 410-sample and 323-sample cohorts, and its interaction with the H:I ratio was evaluated. The authors found that “high MGI was associated with significantly worse outcome only in combination with high HOXB13:IL17BR, and likewise, high HOXB13:IL17BR was significantly associated with poor outcome only in combination with high MGI.” The investigators concluded that “the combination of MGI and HOXB13:IL17BR outperforms either alone and identifies a subgroup (∼30%) of early-stage estrogen receptor-positive breast cancer patients with very poor outcome despite endocrine therapy.32
The BCI has been recently demonstrated to be discerning in other respects. The IDEAL trial attempted to answer the pressing question of the optimal duration of extended endocrine therapy (EET), after completing five years of initial aromatase inhibitor (AI)–based adjuvant therapy.33 The IDEAL trial assessed the ability of BCI (H/I) to predict endocrine benefit of 2.5 vs. five years of extended letrozole using available tumor specimen samples from 908 HR+ patients (73% lymph node-positive, disease free at 2.5 years), with 88% and 68% of which were receiving prior treatment with an aromatase inhibitor (AI) or chemotherapy, respectively. It was found that BCI by H/I status (High vs. Low) was “significantly predictive of response from extended letrozole in the overall (N = 908) and pN+ [lymph node-positive] (N = 664) cohorts”, leading the researchers to exalt the BCI as an important genomic tool for future use.33
Predictor Analysis of Microarray 50
The Predictor Analysis of Microarray 50 (PAM50, by Prosigna) is a 50-gene test that characterizes an individual tumor by intrinsic subtype. It was designed to determine the intrinsic subtype of a cancer using only 50 prespecified genes. The four subtypes are “luminal A, luminal B, HER2-enriched, and basal-like.”34 Results from the PAM50 are used to generate the risk of recurrence (ROR) score, which can stratify patients with ER-positive disease into high, medium, and low subsets. The test can be performed on formalin-fixed, paraffin-embedded tissue with a high degree of analytical validity.9,35
This test was validated by Parker, et al. (2009) The investigators developed this 50-gene set from 189 prototype samples and evaluated 761 patients for prognosis. The four discrete subtypes were found to be prognostically significant and predicted neoadjuvant chemotherapy efficacy with a negative predictive value for pathologic complete response (to a taxane and anthracycline regimen) of 97%. The authors concluded “diagnosis by intrinsic subtype adds significant prognostic and predictive information to standard parameters for patients with breast cancer.”34
Recently, the intrinsic molecular subtype generated by the Prosigna assay has been demonstrated to be useful in guiding the inclusion and exclusion of an anthracycline to CMF-based chemotherapy. The DBCG89D trial randomized 980 high-risk early breast cancer patients to adjuvant CMF (cyclophosphamide, methotrexate and fluorouracil) or CEF (cyclophosphamide, epirubicin and fluorouracil), ultimately using the data from 686 tumor samples. Continuous ROR score was associated with distant recurrence for CMF and CEF (hazard ratios of 1.20 and 1.04, respectively), and distant recurrence was reported to be significantly longer in those patients with HER2-enriched tumors treated with CEF than with CMF. The results suggested that the benefit of CEF as compared to CMF correlate with an increasing ROR score, and so “patients with HER2-enriched breast cancer derived substantial benefit from anthracycline chemotherapy whereas anthracyclines are not an essential component of chemotherapy for patients with luminal subtypes”, and, similarly, “A benefit from substituting methotrexate with epirubicin is observed among patients with HER2-enriched or Basal-like subtypes whereas there is a lack of such benefit among patients with a Luminal A or Luminal B subtypes.”36 However, the authors admit that the results of the study may potentially be skewed by the retrospective evaluation’s use of only a small subset of patients’ data and the lack of optimal instrumentation and testing, since one-third of the patients were ER-positive but endocrine therapies were not employed.36
MammaPrint
MammaPrint is a 70-gene test that assesses the risk of recurrence within the first 5-10 years of diagnosis. The test reports risk in one of two categories, “low” and “high.” MammaPrint’s validation places “low” risk at a 1.3% chance (95% confidence interval: 0-3.1%) that cancer will recur in five years whereas those classified as “high” risk have a 11.7% chance for the cancer to recur in five years (95% confidence interval: 6.6%-16.8%).37,38 Agendia offers other breast cancer assessments, such as BluePrint. BluePrint is an 80-gene molecular subtyping assay, which “analyzes the activity” of its genes. It is proposed to identify a tumor’s molecular “subtype”: luminal A/B, basal, or HER2. Each subtype has varying molecular characteristics (ER-positive or negative, HER2-positive or negative, et al), which is proposed to help clinicians provide targeted treatment for a patient.39
MammaPrint was validated by Cardoso, et al. (2016)The authors determined the risk recurrence of 6693 patients. Of these 6693 patients, 1550 of them were found to have high clinical risk and low genomic risk, and the rate of survival without distant metastasis in this cohort was 94.7% among those not receiving chemotherapy. The difference in survival rate between these patients and those receiving chemotherapy was 1.5%. The authors estimated that “given these findings, approximately 46% of women with breast cancer who are at high clinical risk might not require chemotherapy.”40
However, it may be possible that new prognostic assays are beginning to make their foray. Prat, et al. (2020) derived a prognostic assay that integrates multiple data types for predicting survival outcome in patients with newly diagnosed HER2-positive breast cancer to aid treatment decisions for those with early-stage HER2-positive breast cancer, where escalation or de-escalation of systemic therapy is a point of contention. Using clinical-pathological data on stromal tumor-infiltrating lymphocytes, PAM50 subtypes, and expression of 55 genes obtained from patients who participated in the Short-HER phase 3 trial, Prat, et al. (2020) then evaluated their model based on a combined dataset of 267 patients with early-stage HER2-positive breast cancer treated with different neoadjuvant and adjuvant anti-HER2-based combinations and from four other studies (PAMELA, CHER-LOB, Hospital Clinic, and Padova) with disease-free survival outcome data. The final model, named HER2DX, was comprised of data from 435 (35%) of 1254 patients for tumor size (T1 vs rest), nodal status (N0 vs rest), number of tumor-infiltrating lymphocytes (continuous variable), subtype (HER2-enriched and basal-like vs rest), and 13 genes. The authors exalt the analytical power of HER2DX by reporting that it was “significantly associated with disease-free survival as a continuous variable” and reported that the 5-year disease-free survival in the HER2DX low-risk group was 93.5% (89.0-98.3%) whereas the high-risk group reported 81.1% (71.5-92.1). The researchers assert that HER2DX “identifies patients with early-stage, HER2-positive breast cancer who might be candidates for escalated or de-escalated systemic treatment” and that “Future clinical validation of HER2DX seems warranted to establish its use in different scenarios, especially in the neoadjuvant setting.”41
DCISionRT®
The DCISionRT test out of Prelude Corporation aims to provide patients with three results: 1) their overall score (continuous risk score and low vs elevated category, 2) their rate of recurrence over 10 years, and 3) their benefit from radiation. The intended demographic is females aged 26 and older who have been diagnosed with ductal carcinoma in situ (DCIS) of the breast, and who have been treated with breast conserving surgery or a core biopsy.42
Recognizing that patients diagnosed with DCIS are at risk of recurrence (30%) within 10 years following breast conserving surgery and that election of RT is not always beneficial, the SweDCIS randomized clinical trials also investigated the validity of the DCISionRT biosignature, which combines an evaluation of protein expression (PR, HER2, Ki67, FOXA1, p16/INK4A, SIAH2, COX2) with clinicopathologic factors (age, tumor size, margin status, palpability) and mapped it along a continuous Decision Score (DS) on a scale of 1 to 10 with corresponding 10-year risks after breast conserving surgery with and without radiation therapy.43 The authors found that “using the pre-specified DS threshold of 3.0 to define DS Low and Elevated Risk groups, approximately 50% of women were classified into the Elevated Risk group”, which is significant as many women who would have been erroneously sorted into one risk group based on classical clinicopathology were found to be better reclassified into opposite risk groups based on their DS. Finally, in the DS Elevated Risk group, the addition of radiation therapy was found to reduce absolute 10-year rates for TotBE by 15.9%, and for InvBE by 9.4%, while the DS Low Risk Group boasted 10-year DCIS rates of 0.7% and 5.2%, and InvBE rates of 6.5% and 7.7%, with and without RT, respectively, indicating to the authors that “Radiotherapy benefit was statistically significant in patients with elevated DS but not in patients with low DS by DCISionRT”, though it is also admitted that this evaluation may not reflect modern practice and modern risk profiles because the women participating in the trials were randomly assigned between 1987 and 2000.43
Bremer, et al. (2018) reported on the development and the cross-validation of the DCISionRT biosignature using archived tissue samples from Uppsala University Hospital and Västmanland County Hospital, Sweden (UUH), from 1986 and 2004 and samples from between 1999 and 2008 at the University of Massachusetts, Worcester (UMass), in collaboration with PreludeDx. The study population comprised 721 female patients diagnosed with a primary DCIS and treated with BCS identified from 2 study sites, with mean and median follow-ups of 10 years and 9 years, respectively. Here, the authors found the Decision Score reclassified greater than 50% of the low- and intermediate-grade patients as elevated risk, and 33% of patients that were grade III or had tumor size >2.5 cm were reclassified as low risk. Moreover, it was found that in the low clinicopathological risk group—defined to have tumor size less than 25 mm, low or intermediate grade, and nonpalpable and clear margins—DS reclassified 42% of these patients into the Elevated Risk Group (DS > 3.0). When a linear panel using weighted clinicopathologic and molecular factors to assess recurrence risk was constructed for comparison with the proprietary biosignature, it could not identify a clinically meaningful low-risk group. The researchers reported that “These reclassified patients had substantial 10-year risks, 23% (95% CI: 7%–36%) IBC and 31% (95% CI: 14%–45%) IBE, when not treated with RT” and that while patients in the DS Low Group had similar outcomes regardless of whether they received RT, patients treated with RT and classified into the Elevated Risk Group received a 70% or greater risk reduction than those without RT.44 As such, Bremer, et al. (2018) claims that these “findings reflect the effectiveness of the biological signature to identify patients with favorable tumor biology and separate them from the Elevated Risk Group of patients that have the highest likelihood of recurrence.”44
These results corroborate other investigations into the utility of DCISionRT in guiding treatment management. The prospective, multi-institutional PREDICT study found that DCISionRT testing can lead to great changes in radiation therapy recommendations, such that of the 539 women participating in the study, 69% were initially slated RT pre-test (n = 374) by treating clinicians based on traditional clinicopathology and 46% were recommended to not receive RT post-test (n = 165). But of those initially recommended to not receive RT (n = 165), 35% were then recommended to receive RT post-test (n = 58). As such, the authors reported a significant overall change in the RT recommendation for 42% of women (n = 539, p < 0.001) and a net reduction in the RT recommendation by 20%.45 Furthermore, after DCISionRT testing, elevated versus low DCISionRT score (DS>3 vs. DS ≤ 3) was found to be the strongest factor for an RT recommendation (odds ratio of 43.1) compared against all other factors (age, grade, tumor size, margin status, patient preference). However, the authors acknowledge that there exist limitations to the study, namely that long-term clinical outcomes and satisfaction ratings from patients and physicians are unavailable for analysis and that data for RT recommendations are limited to post-surgery/pre-testing and post-surgery/post-testing time points.45 Thus, while the results seem promising, the real predictive power of the DCISionRT test is still uncertain. Additional information will be critical to evaluating the test’s value and valence—whether positive or negative—in ultimately determining the benefit of adjuvant RT following BCS.
Dabbs, et al. (2023) studied the analytical validity, performance assessment, and clinical performance validation and clinical utility for the DCISionRT test. The authors performed an analytical validation assessment on multiple DCIS molecular assays; the analytical validation test was conducted based on Clinical and Laboratory Standards Institute guidelines. “The analytic validation showed that the molecular assays that are part of DCISionRT test have high sensitivity, specificity, and accuracy/reproducibility (≥95%).” The authors concluded that the DCISionRT has high analytical validity.46
BBDRisk Dx®
The BBDRisk Dx® test is “the first genomic risk test designed specifically for women diagnosed with breast masses that have one or more of the following: Atypical Ductal Hyperplasias (ADH), Atypical Lobular Hyperplasia (ALH), Usual Ductal Hyperplasias (UDH), Papilloma and Sclerosing Adenosis.”47 The test assays four cancer biomarker oncoproteins—Matrix Metalloproteinase-1 (MMP-1), Carcinoembryonic Antigen-related Cell Adhesion Molecule 6 (CEACAM6), Hyaluronoglucosaminidase 1 (HYAL1), Highly Expressed in Cancer Protein 1 (HEC1)—requiring a sample of formalin fixed paraffin-embedded (FFPE) tissue for the immunohistochemistry. The four-marker profile is then rendered to produce a Cancer Risk Score between the range of 0 and 10, classifying the patient into the ‘Low Risk’ Group (Risk Score =< 0.5), the ‘Intermediate Risk’ group (>0.5 but =<5.4), and the ‘High Risk’ group (>5.4). These scores help provide the patient with information about their chances of developing future breast cancer over 5 to 10 years—as compared with women with similar profiles—and guide physician discretion in proposing prophylactic endocrine treatments and or surgery.48
Poola, et al. (2019) explored the effectiveness of the four-marker signature of MMP-1, CEACAM6, HYAL1, and HEC1 in identifying the benefit that high-risk patients presenting atypical and nonatypical hyperplasias would derive from preventive therapies. A total of 440 tissues were included in the study and categorized into two groups: a cancer group of 162 specimens that developed DCIS after a minimum of one year and a maximum of 13 years, and a cancer-free group of 278 specimens from subjects who did not have prior breast cancer and did not develop for a minimum of five years and a maximum of 19 years. A ‘cancer risk score’ (composite value between 0 and 10) was obtained by combining the multiple marker levels through using piecewise linear regression.49 From constructing box plots and density plots of the scores generated from the 440 samples, the authors found that in the risk scores of the cancer-free group peaked around 0.5, while the scores peaked around 9.0 for the cancer group, suggesting that the “risk signature segregates the cancer group from the cancer-free group of tissues.” Moreover, analysis of the risk scores between atypical and nonatypical tissues for any differences using t test, yielded significantly higher values in the cancer group of atypical tissues (P = 0.02) than the nonatypical tissues but the authors did not find any significant differences in the cancer-free groups. Plots produced from Kaplan-Meier survival analysis demonstrated that “In the first 5 years, 53% of subjects in the high-risk group, 12% in the intermediate-risk group, and 1% in the low-risk group developed cancer”, and 73% of atypical subjects in high-risk group developed cancer in the first five years, while in the nonatypical group the cancer rates were only 34%. Meanwhile, mapping cancer risk scores to percent cancer rates in five years, 10 years, 15 years, and beyond 15 years produced a direct and positive correlation.49
Clinical Utility and Validity
Blok, et al. (2018) performed a meta-analysis of the MammaPrint, OncotypeDX, PAM50/Prosigna and Endopredict assays. The authors investigated the evidence available on both the clinical utility and economic value of these assays. Most of the observed studies concluded that genomic profiling contributed to less chemotherapy use. However, the authors caution that “absolute numbers should be interpreted carefully, since some tests are less frequently studied than others” especially as “the clinical consensus on adjuvant chemotherapy is that we are most likely over-treating our patients, since we are not capable of identifying patients that will or will not benefit from chemotherapy using the current clinicopathological parameters.” The authors note that Petkov, et al. (2016) retrospectively matched OncoType DX use with SEER registry data for over 40000 patients and found that “patients with node-negative, HR+, HER2- breast cancer which underwent testing (n = 40,134, 22.7% chemotherapy) had no lower chemotherapy use compared to patients that were not tested (n = 144,056, 22.2% chemotherapy).” The authors noted that 90% of the 44 economic analyses concluded that genomic testing was cost-effective; however, various biases are mentioned (publication bias, publications using overlapping samples, and more).50
Gustavsen, et al. (2014) performed an economic analysis of the Breast Cancer Index and found that “Costs associated with adjuvant chemotherapy, toxicity, follow-up, endocrine therapy, and recurrence were modeled over 10 years. The models examined cost utility compared with standard practice when used at diagnosis and in patients disease-free at 5 years post diagnosis.” The authors calculated BCI to save approximately $3803 per patient for the newly diagnosed population and $1803 per patient in the five years post diagnosis population. These savings were projected to come from “adjuvant chemotherapy use, extended endocrine therapy use, and endocrine therapy compliance.”52
Camp, et al. (2019) investigated a new reinterpretative technique of the PAM50 assay. The authors reorganized the gene expression data provided by the assay into five “quantitative, orthogonal, multigene breast tumor traits” and stated that these “dimensions” better represented breast cancer pedigrees. They re-assessed the data of the GEICAM/9906 clinical trial with these dimensions. After reorganization, the authors concluded that dimensions “PC1 and PC5” were associated with disease-free survival, and low “PC3 and PC4” indicated response to paclitaxel. However, high PC3 or PC4 did not show survival advantage.53
Wuerstlein, et al. (2019) evaluated the clinical utility of MammaPrint and its intended adjunct test, BluePrint. Physicians were asked to provide initial therapy recommendations (based on clinicopathological factors), then were provided results of MammaPrint/BluePrint risk stratifiers. Then the same physicians provided new treatment recommendations, and the subsequent treatment was recorded. The authors identified a switch in treatment in 29.1% of cases. Physician adherence to MammaPrint risk assessment was over 90% in both groups of risk (low and high). Even when physicians and risk stratifiers disagreed, the physicians tended to recommend treatments based on the risk stratifier. Overall, the authors concluded that “MammaPrint and BluePrint test results strongly impacted physicians’ therapy decisions in luminal EBC with up to three involved lymph nodes.”54
Toole, et al. (2014) evaluated the similarities and differences in genetic profiles between primary breast cancers in patients with multiple primaries. The authors also evaluated whether obtaining genetic profiling scores (OncoType DX) on each primary affected chemotherapy decisions. In total, 22 patients had multiple tumor samples sent for analysis, and the authors found that six patients had their chemotherapy recommendations changed based on differing OncoType scores (between their samples). The authors also noted that scores tended to differ more between tumors appearing simultaneously on different breasts compared to multiple tumors on the same breast.55
Martin, et al. (2016) compared the use of EndoPredict® (EPclin) and PAM50 risk of recurrence (ROR) scores in node-positive breast cancer to predict distant metastasis-free survival (MFS). ROR scores can be based using subtype (ROR-S); subtype and proliferation (ROR-P); subtype and tumor size (ROR-T); and subtype, proliferation, and tumor size combined (ROR-PT). They found that “Predictors including clinical information showed superior prognostic performance compared to molecular scores alone (10-year MFS, low-risk group: ROR-T 88 %; ROR-PT 92 %; EPclin 100 %). The EPclin-based risk stratification achieved a significantly improved prediction of MFS compared to ROR-T, but not ROR-PT.”56
Ding, et al. (2019) used the 21-gene recurrence score for patients with pure mucinous breast cancer (PMBC). This multi-year study of 8048 female PMBC patients categorized the patients based on RS risk groups of low, intermediate, and high RS risk groups. They found the distribution to be 64.9%, 31.9%, and 3.2%, respectively. For PMBC patients, the authors note that PMBC patients do show significantly different 5-year survival rates based on ER status. They do conclude that RS can be used with PMBC patients…”age, progesterone receptor status, and grade could independently predict RS.”57 These data support the findings of a considerably smaller comparative study (n=35 PMBC patients and n=70 IDC patients) that show on average PMBC patients score lower than IDC patients (average RS 21.26 and 24.40, respectively); however, they authors do conclude that “patients with high RS in the PMBC group might be recommended to receive adjuvant chemotherapy” since 8.57% of PMBC patients have RS scores within the highest risk stratification.58 Both studies contradict an earlier, smaller study (n=10 TC patients and n=33 MC patients) that reported no patients within the high-risk RS category.59
Kizy, et al. (2018) conducted a large retrospective study of various subtypes of T1-T2N0 estrogen receptor-positive breast cancer (n=83665) was published in 2018. Both mucinous and tubular histologies were included in the study. The authors note considerable differences in RS scores between the various histologies; for example, they found that 1.0% and 28.5% of tubular adenocarcinoma patients had RS scores in the high and intermediate ranges, respectively, whereas mucinous adenocarcinoma patients fared worse with 3.4% and 28.8%, respectively. It should be noted, however, that only 2.1% of the patients included in the study had been diagnosed with mucinous adenocarcinoma and 0.6% with tubular adenocarcinoma.61 Similarly, a study by Kizy et al also reports that 4% of tubular carcinoma patients were classified as high risk based on the Trial Assigning Individualized Options for Treatment RS criteria.60
Harnan, et al. (2019) released an in-depth systemic review of the effectiveness and cost-effectiveness of Oncotype DX®, MammaPrint®, Prosigna®, EndoPredict®, and immunohistochemistry 4 (IHC4). For this review, the authors included a total of 153 studies, including the MINDACT RCT for MammaPrint. The authors note that a limitation of this systemic review, and of the current field in general, is that only one RCT has been completed to date. Their results include the following:
- “There is limited and varying evidence that oncotype DX and MammaPrint can predict benefit from chemotherapy.”
- “The health economic analysis suggests that the incremental cost-effectiveness ratios for the tests versus current practice are broadly favourable for the following scenarios: (1) oncotype DX, for the LN0 subgroup with a Nottingham Prognostic Index (NPI) of > 3.4 and the one to three positive lymph nodes (LN1–3) subgroup (if a predictive benefit is assumed); (2) IHC4 plus clinical factors (IHC4+C), for all patient subgroups; (3) Prosigna, for the LN0 subgroup with a NPI of > 3.4 and the LN1–3 subgroup; (4) EndoPredict Clinical, for the LN1–3 subgroup only [emphasis added]; and (5) MammaPrint, for no subgroups.”62
Qi, et al. (2021) examined the efficacy of the 21-gene assay (Oncotype DX) in core needle biopsies (CNB) as compared to breast cancer resection specimens (the gold standard for obtaining IHC or other molecular information, as they exhibit the most abundant tumor characteristics). The 21-gene assay can be applied to a CNB and CNBs do have the advantage of having a more standardized fixation protocol as compared to resection specimens, which have many different protocols available and often have a greater time between loss of blood supply and the initiation of fixation due to the complexity of surgery. While CNBs are more reliable in determining hormone receptor status due to decreased loss of antigens from the fixation process that resection specimens undergo, CNBs cannot account for crush artefacts which can lead to inaccurate results because of tumor heterogeneity. CNBs are also less accurate than resection specimens in determining the histological characteristics of a tumor, such as tumor invasiveness and tumor grade. It’s also important to note that intratumoral heterogeneity can lead to an underestimation of tumor genomic characteristics because of varying gene expression signations found within different regions of the same tumor.
Qi, et al. (2021) utilized a paired CNB and resection specimen approach to assess the concordance of the Oncotype DX results between the CNB and resection samples. They found that determination of 21-gene assay results from CNBs are reliable, providing results which closely parallel those obtained from resection specimens. However, there is no clear data indicating that tumor grade can be determined using a CNB sample.63
Sennerstam, et al. (2017) conducted retrospective study looking at the long-term outcomes of patients with breast cancer diagnosed with fine-needle aspiration biopsy (FNAB) vs CNB in the 1970s and the 1990s (diagnosis using FNAB from 1971-1976 (n=354); diagnosed using CNB from 1991-1995 (n=1,729)) shows a potentially alarming trend. The authors treatment, mammography screening, tumor size, DNA ploidy, and patient age matched samples from the two decades to get two strictly matched samples representing FNAB (n=181) and CNB (n=203), which were selected for a 15-year follow-up study. When comparing the rates of distant metastasis in the strictly matched patient groups, they found that 5-15 years after diagnosis of the primary tumor, patients diagnosed via CNB had significantly higher rates of metastases as compared to those diagnosed via FNAB.64
American Society of Clinical Oncology
In 2016, ASCO provided recommendations on appropriate use of breast tumor biomarker assay results to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer. ASCO recommends that “in addition to estrogen and progesterone receptors and human epidermal growth factor receptor 2, the panel found sufficient evidence of clinical utility for the biomarker assays Oncotype DX, EndoPredict, PAM50, Breast Cancer Index, and urokinase plasminogen activator and plasminogen activator inhibitor type 1 in specific subgroups of breast cancer.”65
ASCO also made the following recommendations:
- “If a patient has ER/PgR-positive, HER2-negative (node-negative) breast cancer, the clinician may use the 21-gene recurrence score to guide decisions on adjuvant systemic chemotherapy. Type: evidence based. Evidence quality: high. Strength of recommendation: strong.
- If a patient has ER/PgR-positive, HER2-negative (node-positive) breast cancer, the clinician should not use the 21-gene RS to guide decisions on adjuvant systemic chemotherapy. Type: evidence based. Evidence quality: intermediate. Strength of recommendation: moderate.
- If a patient has HER2-positive breast cancer or TN breast cancer, the clinician should not use the 21-gene RS to guide decisions on adjuvant systemic therapy. Type: informal consensus. Evidence quality: insufficient. Strength of recommendation: strong
- If a patient has ER/PgR-positive, HER2-negative (node-positive) breast cancer, the clinician should not use the Breast Cancer Index, the PAM50-ROR, the 12-gene risk score, or the 21-gene RS, (EndoPredict), to guide decisions on adjuvant systemic therapy.
- If a patient has ER/PgR-positive, HER2-negative (node-negative) breast cancer, the clinician may use urokinase plasminogen activator (uPA), plasminogen activator inhibitor type 1 (PAI-1), the Breast Cancer Index, the PAM50 risk of recurrence (ROR) score, the 12-gene risk score (EndoPredict), and the 21-gene recurrence score (Oncotype DX) to guide decisions on adjuvant systemic therapy. If the patient has had 5 years of endocrine therapy without evidence of recurrence, the clinician should not use multiparameter gene expression or protein assays (Oncotype DX, EndoPredict, PAM50, Breast Cancer Index, or IHC4) to guide decisions on extended endocrine therapy.
- If a patient has HER2-positive breast cancer or TN breast cancer, the clinician should not use uPA, IHC4, the 12-gene risk score (EndoPredict), PAI-1, the 21-gene RS (Oncotype DX), the five-protein assay (Mammostrat), the Breast Cancer Index or TILs to guide decisions on adjuvant systemic therapy. The clinician should not use circulating tumor cells (CTCs) to guide decisions on adjuvant systemic therapy.
- If a patient has HER2-positive breast cancer, the clinician should not use the PAM50-ROR to guide decisions on adjuvant systemic therapy. Type: informal consensus. Evidence quality: insufficient. Strength of recommendation: strong
- If a patient has TN breast cancer, the clinician should not use the PAM50-ROR to guide decisions on adjuvant systemic therapy. Type: informal consensus. Evidence quality: insufficient. Strength of recommendation: strong.”65
In 2017 the ASCO,66 based on a review of the MINDACT study publication, revised their guidelines on Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women with Early-Stage Invasive Breast Cancer to state the following:
“Recommendation 1.1.1 (update of Recommendation 1.7).
If a patient has ER/PgR-positive, HER2-negative, node-negative, breast cancer, the MammaPrint assay may be used in those with high clinical risk per MINDACT categorization to inform decisions on withholding adjuvant systemic chemotherapy due to its ability to identify a good prognosis population with potentially limited chemotherapy benefit (Type: evidence based; Evidence quality: high; Strength of recommendation: strong).
Recommendation 1.1.2 (update of Recommendation 1.7).
If a patient has ER/PgR-positive, HER2-negative, node-negative, breast cancer, the MammaPrint assay should not be used in those with low clinical risk per MINDACT categorization to inform decisions on withholding adjuvant systemic chemotherapy as women in the low clinical risk category had excellent outcomes and did not appear to benefit from chemotherapy even with a genomic high-risk cancer (Type: evidence based; Evidence quality: high; Strength of recommendation: strong).”
Recommendation 1.2.2: (update of 2016 recommendation 1.7): If a patient has ER/PgR-positive, HER2-negative, node-positive, breast cancer, the MammaPrint assay should not be used in patients with one to three positive nodes and at low clinical risk per MINDACT categorization to inform decisions on withholding adjuvant systemic chemotherapy. There are insufficient data on the clinical utility of MammaPrint in this specific patient population (Type: informal consensus; Evidence quality: low; Strength of recommendation: moderate).
Recommendation 1.3: (update of 2016 recommendation 1.8): If a patient has HER2-positive breast cancer, the clinician should not use the MammaPrint assay to guide decisions on adjuvant systemic therapy. Additional studies are required to address the role of MammaPrint in patients with this tumor subtype who are also receiving HER2-targeted therapy (Type: informal consensus; Evidence quality: low; Strength of recommendation: moderate).
Recommendation 1.4: (update of 2016 recommendation 1.9): If a patient has ER/PgR negative and HER2-negative (triple negative) breast cancer, the clinician should not use the MammaPrint assay to guide decisions on adjuvant systemic chemotherapy (Type: informal consensus; Evidence quality: insufficient; Strength of recommendation: strong).”66
ASCO published an update regarding the “Role of Patient and Disease Factors in Adjuvant Systemic Therapy Decision Making for Early-Stage, Operable Breast Cancer”, which was released in response to the results of two-phase III trials, MINDACT and TAILORx. The updated guidelines state that the clinical trials found evidence of clinical utility for both node-positive and node-negative patients, but only for patients determined to be at high clinical risk. Therefore, the panel did not recommend use of MammaPrint for patients determined to be of low clinical risk.67
In a focused update published in 2019, the ASCO addressed the use of Oncotype DX in the following recommendation:
“For patients with hormone receptor-positive, axillary node-negative breast cancer whose tumors have Oncotype DX recurrence scores of less than 26, there is little to no benefit from chemotherapy, especially for patients older than age 50 years. Clinicians may recommend endocrine therapy alone for women older than age 50 years. For patients 50 years of age or younger with recurrence scores of 16 to 25, clinicians may offer chemoendocrine therapy. Patients with recurrence scores greater than 30 should be considered candidates for chemoendocrine therapy. Based on informal consensus, the panel recommends that oncologists may offer chemoendocrine therapy to these patients with recurrence scores of 26 to 30.”68
In 2022, the ASCO released a guideline update on biomarkers for adjuvant endocrine and chemotherapy in early-stage breast cancer. The ASCO recommends that
- “Recommendation 1.1. If a patient has node-negative breast cancer, the clinician may use the Oncotype DX test to guide decisions for adjuvant endocrine and chemotherapy (Type: evidence-based; Evidence quality: high; Strength of recommendation: strong).
- Recommendation 1.2. In the group of patients in Recommendation 1.1 with Oncotype DX recurrence score greater or equal to 26, the clinician should offer chemoendocrine therapy (Type: evidence-based; Evidence quality: high; Strength of recommendation: strong).
- Recommendation 1.3. In the group of patients in Recommendation 1.1 who are 50 years of age or younger with Oncotype DX recurrence score 16 to 25, the clinician may offer chemoendocrine therapy (Type: evidence-based; Evidence quality: intermediate; Strength of recommendation: moderate).
- Recommendation 1.4. If a patient is postmenopausal and has node-positive breast cancer with 1-3 positive nodes, the clinician may use the Oncotype DX test to guide decisions for adjuvant endocrine and chemotherapy (Type: evidence-based; Evidence quality: high; Strength of recommendation: strong).
- Recommendation 1.5. In the group of patients in Recommendation 1.4, the clinician should offer chemoendocrine therapy for those whose Oncotype DX recurrence score is ≥ 26 (Type: evidence-based; Evidence quality: high; Strength of recommendation: strong).
- Recommendation 1.6. If a patient is premenopausal and has node-positive breast cancer with 1-3 positive nodes, Oncotype DX test should not be offered to guide decisions for adjuvant systemic chemotherapy (Type: evidence-based; Evidence quality: high; Strength of recommendation: moderate).
- Recommendation 1.7. If a patient has node-positive breast cancer with ≥ 4 positive nodes, the evidence on the clinical utility of routine Oncotype DX to guide decisions for adjuvant endocrine and chemotherapy is insufficient to recommend its use (Type: informal consensus; Evidence quality: insufficient; Strength of recommendation: moderate).”69
American Cancer Society (ACS)
In 2017 the American Cancer Society (ACS) released a revision to the American Joint Cancer Committee’s Eighth Edition Cancer Staging Manual. The revision suggests that:
“For patients with hormone receptor-positive, HER2-negative, and lymph node-negative tumors, a21-gene (Oncotype Dx) recurrence score less than 11, regardless of T size, places the tumor into the same prognostic category as T1a-T1b N0 M0, and the tumor is staged using the AJCC prognostic stage group table as stage I.
For patients with hormone receptor-positive, HER2-negative, and lymph node-negative tumors, a MammaPrint low-risk score, regardless of T size, places the tumor into the same prognostic category as T1a-T1b N0 M0.
For patients with hormone receptor-positive, HER2-negative, and lymph node-negative tumors, a12-gene (EndoPredict) low-risk score, regardless of T size, places the tumor into the same prognostic category as T1a-T1b N0 M0.
For patients with hormone receptor-positive, HER2-negative, and lymph node-negative tumors, aPAM50 risk–of-recurrence score in the low range, regardless of T size, places the tumor into the same prognostic category as T1a-T1b N0 M0.
For patients with hormone receptor-positive, HER2-negative, and lymph node-negative tumors, a Breast Cancer Index in the low-risk range, regardless of T size, places the tumor into the same prognostic category as T1a-T1b N0 M0.”70
National Comprehensive Cancer Network (NCCN)
The NCCN Clinical Practice Guidelines for Breast Cancer consider five gene expression assays for adjuvant systemic therapy. The five are comprised of the 21-gene Oncotype Dx, the 70-gene MammaPrint, the 50-gene PAM50 (Prosigna), the 12-gene EndoPredict, and the Breast Cancer Index (BCI). All five gene expression assays achieved the NCCN Category of Evidence and Consensus grade of 2A (consensus of appropriateness of intervention based on lower-level evidence) or higher, with Oncotype Dx achieving grade 1 (consensus based on higher-level evidence) for predictive and prognostic use, and MammaPrint achieving grade 1 for prognostic use in patients who are diagnosed as node-negative or with 1 to 3 positive nodes. It should be noted that NCCN identifies only the Oncotype Dx as suitable for prediction of chemotherapy benefit; NCCN states that “Other prognostic gene assays can provide prognostic information but the ability to predict chemotherapy benefit is unknown.” BCI is identified as suitable for predicting benefit of extended adjuvant endocrine therapy.2
The NCCN panel “notes that the multigene assays provide prognostic and therapy-predictive information that complements TNM and biomarker information.” Below is the table from their recommendations and guidelines based on risk scores and nodal statuses of the multigene assays:2
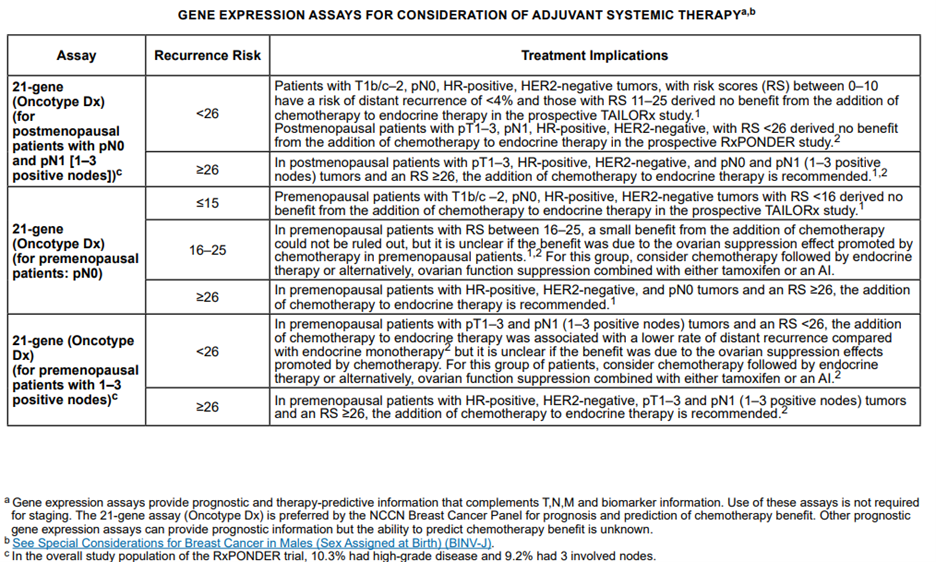
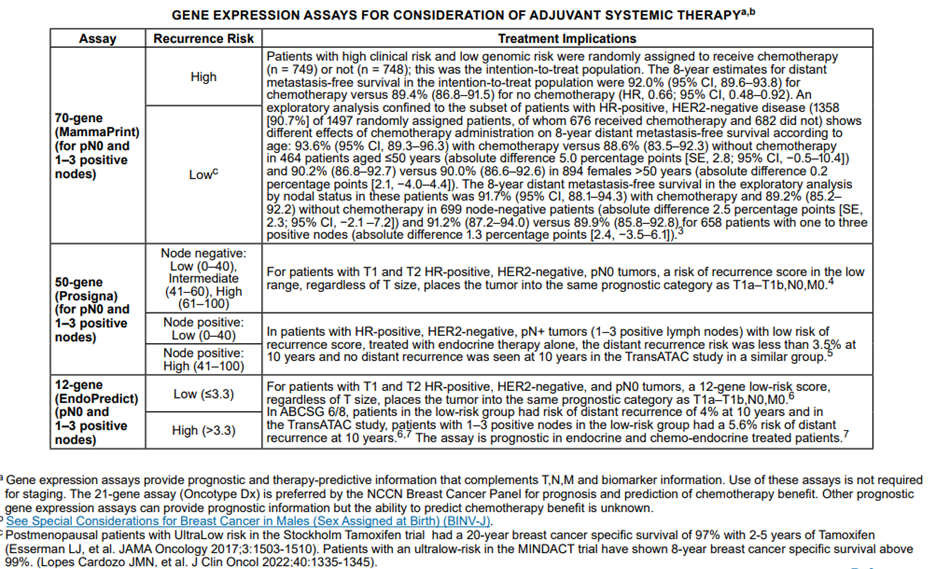
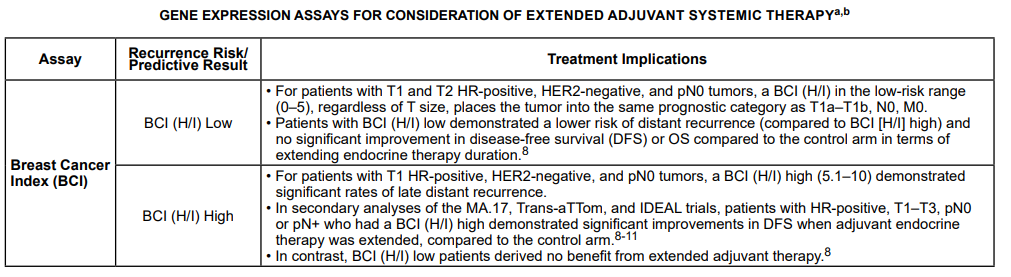

The NCCN also had the following to summarize about the assays mentioned above and their respective interpretations and/or rationales:
For Oncotype DX in node-negative, HR-positive, HER2-negative disease: “Among patients with T1b/c and T2, lymph node-negative, HR-positive, HER2-negative tumors with RS between 0-10, the risk of distant recurrence is low and these patients derive no incremental benefit from the addition of adjuvant chemotherapy to endocrine therapy. At the other end of the spectrum, patients with lymph node-negative, HR-positive, HER2-negative cancers with high RS (≥31) have a higher risk of distant recurrence and secondary analyses of prospective studies demonstrate a clear benefit from adjuvant chemotherapy.”
Further, “for those with intermediate RS (11-25), the recently reported TAILORx trial of postmenopausal patients (n=6711) with lymph node-negative, HR-positive, HER-2 negative breast cancer, showed similar disease-free survival rates at 9-years in those who received adjuvant chemotherapy followed by endocrine therapy compared with endocrine therapy alone.”
Based on secondary analyses of studies involving the use of Oncotype DX, the NCCN states that for individuals with node-positive, HR-positive, HER2-negative disease, “results suggest that in patients with limited nodal disease (1-3 positive lymph nodes) and a low RS, the absolute benefit from chemotherapy is likely to be very small.” On the other hand, “There is a clear benefit from adjuvant chemotherapy in patients with node-positive, HR-positive, HER2-negative tumors, if the RS is high (≥31).” However, “The absolute benefit of chemotherapy in patients with limited lymph node involvement and a RS ≤25 remains to be determined.”
For MammaPrint: The NCCN claims that the “data suggest that the additional benefit of adjuvant chemotherapy in patients with high-clinical risk/low genomic risk is likely to be small” and that the data suggest that “the results of the 70-gene signature do not provide evidence for making recommendations regarding chemotherapy for patients at low clinical risk.”
For PAM50: for “patients with T1 and T2, HR-positive, HER2-negative, lymph node-negative tumors, a ROR in the low range, regardless of tumor size, places the individual into the same prognostic category as T1a-T1b, N0, M0 tumors.
In patients with 1-3 lymph-node positive, HR-positive, HER2-negative disease with low-risk of recurrence score, the distant recurrence risk was less than 3.5% at 10 years with endocrine therapy alone.”
For EndoPredict: “patients with HR-positive, HER-2 negative, and lymph-node node-negative disease with a low-risk score by the 12-gene assay had risk of distant recurrence of 4% at 10 years” and “patients with T1 and T2 HR-positive, HER2-negative, and lymph node-negative tumors, a 12-gene low risk score, regardless of T size, places the tumor into the same prognostic category as T1a-T1b, N0, M0.” Citing another study, the NCCN states the following for EndoPredict: “patients with 1-3 positive nodes in the low-risk group had a 5.6% risk of distant recurrence at 10 years, suggesting that chemotherapy would be of limited benefit in these patients.”
For the BCI: “For patients with T1 and T2 HR-positive, HER2-negative, and lymph node-negative tumors, a BCI in the low-risk range, regardless of T size, places the tumor into the same prognostic category as T1a-T1b, N0, M0. There are limited data as to the role of BCI in HR-positive, HER2-negative, and lymph node-positive breast cancer.”2
The NCCN strongly recommends (Category 1) considering the use of the 21-gene reverse transcriptase polymerase chain reaction (RT-PCR) assay for determining the use of adjuvant chemotherapy in premenopausal and postmenopausal patients with the following tumor characteristics:
- Hormone receptor-positive;
- HER2 [human epidermal growth factor receptor 2]-negative;
- Ductal/NST, lobular, mixed or micropapillary histology;
- pT1, pT2 or pT3 stage; and pN0;
- Tumor >0.5 cm.
The assay may also be considered for pre- and postmenopausal, node-positive (up to three positive nodes) individuals if such individuals are candidates for chemotherapy.
Within a section on special considerations for breast cancer in men, the NCCN states (Category 2A), “Data are limited regarding the use of molecular assays to assess prognosis and to predict benefit from chemotherapy in males with breast cancer. Available data suggest 21-gene assay recurrence score provides prognostic information in males with breast cancer.”2
The NCCN also notes that ER status should be determined for DCIS patients, but do not mention any gene expression tests for evaluation.2
National Institute for Health and Care Excellence (NICE)
Three new guidelines were released by NICE regarding gene expression tests, which are as follows:
1. “EndoPredict (EPclin score), Oncotype DX Breast Recurrence Score and Prosigna are recommended as options for guiding adjuvant chemotherapy decisions for people with oestrogen receptor (ER)-positive, human epidermal growth factor receptor 2 (HER2)-negative and lymph node (LN)-negative (including micrometastatic disease; see section 5.4) early breast cancer, only if:
- they have an intermediate risk of distant recurrence using a validated tool such as PREDICT or the Nottingham Prognostic Index
- the companies provide the tests to the NHS with the discounts agreed in the access proposals
- clinicians and companies make timely, complete and linkable record-level test data available to the National Cancer Registration and Analysis Service as described in the data collection arrangements agreed with NICE (see section 5.29).”
2. “MammaPrint and IHC4+C is not recommended for guiding adjuvant chemotherapy decisions for people with ER- or PR- positive, HER2‑negative and LN-negative early breast cancer.”71
European Society for Medical Oncology (ESMO)
In their guideline ESMO notes that “In cases of uncertainty regarding indications for adjuvant chemotherapy (after consideration of other tests), gene expression assays, such as MammaPrint, Oncotype DX, Prosigna and EndoPredict, may be used, where available. These assays can determine the individual’s recurrence risk as well as potentially predict the benefit of chemotherapy.” This point was unchanged in the 2019 update.”72,73
In a 2019 update, ESMO elaborates on gene expression profiles for breast cancer. They state that “Gene expression profiles, such as MammaPrint, Oncotype DX Recurrence Score, Prosigna and Endopredict, may be used to gain additional prognostic and/or predictive information to complement pathology assessment and to predict the benefit of adjuvant chemotherapy.” They also remark that Prosigna, Endopredict, and Oncotype are intended for ER-positive early breast cancer only and that MammaPrint and OncoType are still being evaluated for clinical utility.73
References:
1. Steiling K, Christenson S. Tools for genetics and genomics: Gene expression profiling. Updated October 3, 2023. https://www.uptodate.com/contents/tools-for-genetics-and-genomics-gene-expression-profiling
2. NCCN. NCCN Clinical Practice Guidelines in Oncology: Breast Cancer v6.2024. NCCN. Updated November 11, 2024. https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: A Cancer Journal for Clinicians. 2019/01/01 2019;69(1):7-34. doi:10.3322/caac.21551
4. Taghian A, Merajver SD. Overview of the treatment of newly diagnosed, non-metastatic breast cancer. Updated January 25. https://www.uptodate.com/contents/overview-of-the-treatment-of-newly-diagnosed-invasive-non-metastatic-breast-cancer
5. Peto R, Davies C, Godwin J, et al. Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet (London, England). Feb 04 2012;379(9814):432-44. doi:10.1016/s0140-6736(11)61625-5
6. Davies C, Godwin J, Gray R, et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet (London, England). Aug 27 2011;378(9793):771-84. doi:10.1016/s0140-6736(11)60993-8
7. Darby S, McGale P, Correa C, et al. Effect of radiotherapy after breast-conserving surgery on 10-year recurrence and 15-year breast cancer death: meta-analysis of individual patient data for 10,801 women in 17 randomised trials. Lancet (London, England). Nov 12 2011;378(9804):1707-16. doi:10.1016/s0140-6736(11)61629-2
8. Forouzanfar MH, Foreman KJ, Delossantos AM, et al. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet (London, England). Oct 22 2011;378(9801):1461-84. doi:10.1016/s0140-6736(11)61351-2
9. Foukakis T, Bergh J. Prognostic and predictive factors in early, nonmetastatic breast cancer. Updated Septmeber 09, 2024. https://www.uptodate.com/contents/prognostic-and-predictive-factors-in-early-non-metastatic-breast-cancer
10. Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. Journal of the National Cancer Institute. Nov 04 2009;101(21):1446-52. doi:10.1093/jnci/djp335
11. Chen R, Xia L, Tu K, et al. Longitudinal personal DNA methylome dynamics in a human with a chronic condition. Nature medicine. Dec 2018;24(12):1930-1939. doi:10.1038/s41591-018-0237-x
12. Rueda OM, Sammut S-J, Seoane JA, et al. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature. 2019/03/01 2019;567(7748):399-404. doi:10.1038/s41586-019-1007-8
13. Savci-Heijink CD, Halfwerk H, Koster J, Horlings HM, van de Vijver MJ. A specific gene expression signature for visceral organ metastasis in breast cancer. BMC Cancer. Apr 8 2019;19(1):333. doi:10.1186/s12885-019-5554-z
14. Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. The New England journal of medicine. Dec 30 2004;351(27):2817-26. doi:10.1056/NEJMoa041588
15. Sparano JA, Gray RJ, Makower DF, et al. Prospective Validation of a 21-Gene Expression Assay in Breast Cancer. The New England journal of medicine. Nov 19 2015;373(21):2005-14. doi:10.1056/NEJMoa1510764
16. Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. Aug 10 2006;24(23):3726-34. doi:10.1200/jco.2005.04.7985
17. Mamounas EP, Tang G, Fisher B, et al. Association between the 21-gene recurrence score assay and risk of locoregional recurrence in node-negative, estrogen receptor-positive breast cancer: results from NSABP B-14 and NSABP B-20. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. Apr 01 2010;28(10):1677-83. doi:10.1200/jco.2009.23.7610
18. Sparano JA, Gray RJ, Makower DF, et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. The New England journal of medicine. Jun 3 2018;doi:10.1056/NEJMoa1804710
19. Kalinsky K, Barlow WE, Meric-Bernstam F, et al. Abstract GS3-00: First results from a phase III randomized clinical trial of standard adjuvant endocrine therapy (ET) +/- chemotherapy (CT) in patients (pts) with 1-3 positive nodes, hormone receptor-positive (HR+) and HER2-negative (HER2-) breast cancer (BC) with recurrence score (RS) < 25: SWOG S1007 (RxPonder). Cancer Research. 2021;81(4 Supplement):GS3-00-GS3-00. doi:10.1158/1538-7445.Sabcs20-gs3-00
20. Lashen A, Toss MS, Fadhil W, Oni G, Madhusudan S, Rakha E. Evaluation oncotype DX(®) 21-gene recurrence score and clinicopathological parameters: a single institutional experience. Histopathology. Apr 2023;82(5):755-766. doi:10.1111/his.14863
21. Myriad. EndoPredict Executive Summary. Myriad Genetics, Inc. Accessed 01/21/2020, 2020. https://myriad.com/managed-care/endopredict/
22. Warf MB, Rajamani S, Krappmann K, et al. Analytical validation of a 12-gene molecular test for the prediction of distant recurrence in breast cancer. Future Science OA. 2017/08/01 2017;3(3):FSO221. doi:10.4155/fsoa-2017-0051
23. Myriad. Myriad EndoPredict® Technical Specifications. Updated 12/10/2019. Accessed 01/27/2020, 2020. https://myriad-library.s3.amazonaws.com/technical-specifications/EndoPredict-Technical-Specifications.pdf
24. Filipits M, Dubsky P, Rudas M, et al. Prediction of Distant Recurrence using EndoPredict among Women with ER+, HER2- Node-Positive and Node-Negative Breast Cancer Treated with Endocrine Therapy Only. Clinical Cancer Research. 2019:clincanres.0376.2019. doi:10.1158/1078-0432.CCR-19-0376
25. Filipits M, Rudas M, Jakesz R, et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clinical cancer research : an official journal of the American Association for Cancer Research. Sep 15 2011;17(18):6012-20. doi:10.1158/1078-0432.Ccr-11-0926
26. Myriad. WHAT IS ENDOPREDICT? https://endopredict.com/overview/
27. Sestak I, Buus R, Cuzick J, et al. Comparison of the Performance of 6 Prognostic Signatures for Estrogen Receptor-Positive Breast Cancer: A Secondary Analysis of a Randomized Clinical Trial. JAMA Oncol. Apr 1 2018;4(4):545-553. doi:10.1001/jamaoncol.2017.5524
28. Sestak I, Martin M, Dubsky P, et al. Prediction of chemotherapy benefit by EndoPredict in patients with breast cancer who received adjuvant endocrine therapy plus chemotherapy or endocrine therapy alone. Breast Cancer Res Treat. Jul 2019;176(2):377-386. doi:10.1007/s10549-019-05226-8
29. Villarreal-Garza C, Lopez-Martinez E, Deneken Z, et al. Change in therapeutic management after the EndoPredict assay in a prospective decision impact study of Mexican premenopausal breast cancer patients. PLOS ONE. 03/11 2020;15:e0228884. doi:10.1371/journal.pone.0228884
30. Dubsky PC, Singer CF, Egle D, et al. The EndoPredict score predicts response to neoadjuvant chemotherapy and neoendocrine therapy in hormone receptor-positive, human epidermal growth factor receptor 2-negative breast cancer patients from the ABCSG-34 trial. Eur J Cancer. Jul 2020;134:99-106. doi:10.1016/j.ejca.2020.04.020
31. Biotheranostics. About Breast Cancer Index (BCI). https://www.breastcancerindex.com/about-breast-cancer-index#
32. Ma X-J, Salunga R, Dahiya S, et al. A Five-Gene Molecular Grade Index and <em>HOXB13:IL17BR</em> Are Complementary Prognostic Factors in Early Stage Breast Cancer. Clinical Cancer Research. 2008;14(9):2601. doi:10.1158/1078-0432.CCR-07-5026
33. Liefers G-J, Noordhoek I, Treuner K, et al. Breast cancer index (BCI) predicts benefit of two-and-a-half versus five years of extended endocrine therapy in HR+ breast cancer patients treated in the ideal trial. Journal of Clinical Oncology. 2020;38(15_suppl):512-512. doi:10.1200/JCO.2020.38.15_suppl.512
34. Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. Mar 10 2009;27(8):1160-7. doi:10.1200/jco.2008.18.1370
35. Nielsen TO, Parker JS, Leung S, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. Nov 01 2010;16(21):5222-32. doi:10.1158/1078-0432.ccr-10-1282
36. Jensen M-B, Lænkholm A-V, Balslev E, et al. The Prosigna 50-gene profile and responsiveness to adjuvant anthracycline-based chemotherapy in high-risk breast cancer patients. npj Breast Cancer. 2020/02/26 2020;6(1):7. doi:10.1038/s41523-020-0148-0
37. Agendia. MammaPrint: a test to predict risk of breast cancer recurrence. https://agendia.com/mammaprint/
38. Agendia. MammaPrint® FFPE Result. https://www.agendia.com/pdf/high-risk.pdf
39. Agendia. BluePrint: a test to predict likely tumor behavior. https://agendia.com/blueprint/
40. Cardoso F, van’t Veer LJ, Bogaerts J, et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. New England Journal of Medicine. 2016/08/25 2016;375(8):717-729. doi:10.1056/NEJMoa1602253
41. Prat A, Guarneri V, Paré L, et al. A multivariable prognostic score to guide systemic therapy in early-stage HER2-positive breast cancer: a retrospective study with an external evaluation. Lancet Oncol. Nov 2020;21(11):1455-1464. doi:10.1016/s1470-2045(20)30450-2
42. PreludeDx. DCISionRT(R): Welcome to a New Paradigm for DCIS Management. Prelude Corporation. https://preludedx.com/physicians/
43. Wärnberg F, Karlsson P, Holmberg E, et al. Prognostic Risk Assessment and Prediction of Radiotherapy Benefit for Women with Ductal Carcinoma In Situ (DCIS) of the Breast, in a Randomized Clinical Trial (SweDCIS). Cancers. 2021;13(23):6103. doi:10.3390/cancers13236103
44. Bremer T, Whitworth PW, Patel R, et al. A Biological Signature for Breast Ductal Carcinoma In Situ to Predict Radiotherapy Benefit and Assess Recurrence Risk. Clinical cancer research : an official journal of the American Association for Cancer Research. Dec 1 2018;24(23):5895-5901. doi:10.1158/1078-0432.Ccr-18-0842
45. Shah C, Bremer T, Cox C, et al. The Clinical Utility of DCISionRT(®) on Radiation Therapy Decision Making in Patients with Ductal Carcinoma In Situ Following Breast-Conserving Surgery. Ann Surg Oncol. Oct 2021;28(11):5974-5984. doi:10.1245/s10434-021-09903-1
46. Dabbs D, Mittal K, Heineman S, et al. Analytical validation of the 7-gene biosignature for prediction of recurrence risk and radiation therapy benefit for breast ductal carcinoma in situ. Front Oncol. 2023;13:1069059. doi:10.3389/fonc.2023.1069059
47. BBDRiskDx. What is the BBDRISK DX® Test? https://bbdrisk.com/patients/bbdrisk-dx-test/bbdrisk-dx/
48. DESCRIPTION OF THE BBDRisk Dx® TEST. Silbiotech, Inc.; 2021. https://bbdrisk.com/wp-content/uploads/2021/07/downloadable-information-to-give-to-physician.pdf
49. Poola I, Yue Q, Gillespie JW, et al. Breast Hyperplasias, Risk Signature, and Breast Cancer. Cancer Prev Res (Phila). Jul 2019;12(7):471-480. doi:10.1158/1940-6207.Capr-19-0051
50. Blok EJ, Bastiaannet E, van den Hout WB, et al. Systematic review of the clinical and economic value of gene expression profiles for invasive early breast cancer available in Europe. Cancer treatment reviews. Jan 2018;62:74-90. doi:10.1016/j.ctrv.2017.10.012
51. Petkov VI, Miller DP, Howlader N, et al. Breast-cancer-specific mortality in patients treated based on the 21-gene assay: a SEER population-based study. NPJ Breast Cancer. 2016;2:16017. doi:10.1038/npjbcancer.2016.17
52. Gustavsen G, Schroeder B, Kennedy P, et al. Health economic analysis of Breast Cancer Index in patients with ER+, LN- breast cancer. The American journal of managed care. Aug 1 2014;20(8):e302-10.
53. Camp NJ, Madsen MJ, Herranz J, et al. Re-interpretation of PAM50 gene expression as quantitative tumor dimensions shows utility for clinical trials: application to prognosis and response to paclitaxel in breast cancer. Breast Cancer Res Treat. Jan 23 2019;doi:10.1007/s10549-018-05097-5
54. Wuerstlein R, Kates R, Gluz O, et al. Strong impact of MammaPrint and BluePrint on treatment decisions in luminal early breast cancer: results of the WSG-PRIMe study. Breast Cancer Res Treat. Jun 2019;175(2):389-399. doi:10.1007/s10549-018-05075-x
55. Toole MJ, Kidwell KM, Van Poznak C. Oncotype dx results in multiple primary breast cancers. Breast cancer : basic and clinical research. Jan 9 2014;8:1-6. doi:10.4137/bcbcr.S13727
56. Martin M, Brase JC, Ruiz A, et al. Prognostic ability of EndoPredict compared to research-based versions of the PAM50 risk of recurrence (ROR) scores in node-positive, estrogen receptor-positive, and HER2-negative breast cancer. A GEICAM/9906 sub-study. Breast Cancer Res Treat. Feb 2016;156(1):81-9. doi:10.1007/s10549-016-3725-z
57. Ding S, Wu J, Lin C, et al. Predictors for Survival and Distribution of 21-Gene Recurrence Score in Patients With Pure Mucinous Breast Cancer: A SEER Population-Based Retrospective Analysis. Clinical breast cancer. Feb 2019;19(1):e66-e73. doi:10.1016/j.clbc.2018.10.001
58. Wang W, Chen X, Lin L, et al. Distribution and Clinical Utility of the 21-gene Recurrence Score in Pure Mucinous Breast Cancer Patients: a case-control study. Journal of Cancer. 2018;9(18):3216-3224. doi:10.7150/jca.27291
59. Turashvili G, Brogi E, Morrow M, et al. The 21-gene recurrence score in special histologic subtypes of breast cancer with favorable prognosis. Breast Cancer Res Treat. Aug 2017;165(1):65-76. doi:10.1007/s10549-017-4326-1
60. Kizy S, Huang JL, Marmor S, et al. Distribution of 21-Gene Recurrence Scores Among Breast Cancer Histologic Subtypes. Archives of pathology & laboratory medicine. Jun 2018;142(6):735-741. doi:10.5858/arpa.2017-0169-OA
61. Wang J, He ZY, Dong Y, Sun JY, Zhang WW, Wu SG. The Distribution and Outcomes of the 21-Gene Recurrence Score in T1-T2N0 Estrogen Receptor-Positive Breast Cancer With Different Histologic Subtypes. Frontiers in genetics. 2018;9:638. doi:10.3389/fgene.2018.00638
62. Harnan S, Tappenden P, Cooper K, et al. Tumour profiling tests to guide adjuvant chemotherapy decisions in early breast cancer: a systematic review and economic analysis. Health Technol Assess. Jun 2019;23(30):1-328. doi:10.3310/hta23300
63. Qi P, Yang Y, Bai QM, et al. Concordance of the 21-gene assay between core needle biopsy and resection specimens in early breast cancer patients. Breast Cancer Res Treat. Apr 2021;186(2):327-342. doi:10.1007/s10549-020-06075-6
64. Sennerstam RB, Franzen BSH, Wiksell HOT, Auer GU. Core-needle biopsy of breast cancer is associated with a higher rate of distant metastases 5 to 15 years after diagnosis than FNA biopsy. Cancer Cytopathol. Oct 2017;125(10):748-756. doi:10.1002/cncy.21909
65. Harris LN, Ismaila N, McShane LM, et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. Apr 01 2016;34(10):1134-50. doi:10.1200/jco.2015.65.2289
66. Krop I, Ismaila N, Andre F, et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline Focused Update. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. Jul 10 2017;doi:10.1200/jco.2017.74.0472
67. Henry NL, Somerfield MR, Abramson VG, et al. Role of Patient and Disease Factors in Adjuvant Systemic Therapy Decision Making for Early-Stage, Operable Breast Cancer: Update of the ASCO Endorsement of the Cancer Care Ontario Guideline. Journal of Clinical Oncology. 2019/08/01 2019;37(22):1965-1977. doi:10.1200/JCO.19.00948
68. Andre F, Ismaila N, Henry NL, et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: ASCO Clinical Practice Guideline Update—Integration of Results From TAILORx. Journal of Clinical Oncology. 2019/08/01 2019;37(22):1956-1964. doi:10.1200/JCO.19.00945
69. Andre F, Ismaila N, Allison KH, et al. Biomarkers for Adjuvant Endocrine and Chemotherapy in Early-Stage Breast Cancer: ASCO Guideline Update. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. Jun 1 2022;40(16):1816-1837. doi:10.1200/jco.22.00069
70. Giuliano AE, Connolly JL, Edge SB, et al. Breast Cancer-Major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA: A Cancer Journal for Clinicians. 2017;67(4):290-303. doi:10.3322/caac.21393
71. NICE. Tumour profiling tests to guide adjuvant chemotherapy decisions in early breast cancer. https://www.nice.org.uk/guidance/dg58
72. Senkus E, Kyriakides S, Ohno S, et al. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of oncology : official journal of the European Society for Medical Oncology. Sep 2015;26 Suppl 5:v8-30. doi:10.1093/annonc/mdv298
73. Cardoso F, Kyriakides S, Ohno S, et al. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-updagger. Annals of oncology : official journal of the European Society for Medical Oncology. Aug 1 2019;30(8):1194-1220. doi:10.1093/annonc/mdz173
74. FDA. MammaPrint®: 510(k) Substantial Equivalence Determination Decision Summary. 2007. June 22, 2007. https://www.accessdata.fda.gov/cdrh_docs/reviews/K070675.pdf
75. FDA. MammaPrint®: 510(k) Substantial Equivalence Determination Decision Summary. 2009. December 11, 2009. https://www.accessdata.fda.gov/cdrh_docs/reviews/K081092.pdf
76. FDA. MammaPrint® FFPE: 510(k) Substantial Equivalence Determination Decision Summary Assay and Instrument Combination Template. 2015. January 23, 2015. https://www.accessdata.fda.gov/cdrh_docs/reviews/K141142.pdf
77. FDA. Prosigna(TM) Breast Cancer Prognostic Gene Signature Assay: 510(k) Substantial Equivalence Determination Decision Summary Assay and Instrument Combination Template. 2013. 09/06/2013. https://www.accessdata.fda.gov/cdrh_docs/reviews/K130010.pdf
Coding Section
| CPT |
Code Description |
| 81479 |
Unlisted molecular pathology procedure |
| 81518 |
Oncology (breast), mRNA, gene expression profiling by real-time RT-PCR of 11 genes (7 content and 4 housekeeping), utilizing formalin-fixed paraffin-embedded tissue, algorithms reported as percentage risk for metastatic recurrence and likelihood of benefit from extended endocrine therapy |
| 81519 |
Oncology (breast), mRNA, gene expression profiling by real-time RT-PCR of 21 genes, utilizing formalin-fixed paraffin-embedded tissue, algorithm reported as recurrence score |
| 81520 |
Oncology (breast), mrna gene expression profiling by hybrid capture of 58 genes (50 content and 8 housekeeping), utilizing formalin-fixed paraffin-embedded tissue, algorithm reported as a recurrence risk score |
| 81521 |
Oncology (breast), mrna, microarray gene expression profiling of 70 content genes and 465 housekeeping genes, utilizing fresh frozen or formalin-fixed paraffin-embedded tissue, algorithm reported as index related to risk of distant metastasis |
| 81522 |
Oncology (breast), mRNA, gene expression profiling by RT-PCR of 12 genes (8 content and 4 housekeeping), utilizing formalin-fixed paraffin-embedded tissue, algorithm reported as recurrence risk score |
| 81523 |
Oncology (breast), mRNA, next-generation sequencing gene expression profiling of 70 content genes and 31 housekeeping genes, utilizing formalin-fixed paraffin-embedded tissue, algorithm reported as index related to risk to distant metastasis Proprietary test: MammaPrint® Lab/Manufacturer: Agendia, Inc |
| 81599 |
Unlisted multianalyte assay with algorithmic analysis |
| 88360 |
Morphometric analysis, tumor immunohistochemistry (e.g., Her-2/neu, estrogen receptor/progesterone receptor), quantitative or semiquantitative, per specimen, each single antibody stain procedure; manual |
| 88361 |
Morphometric analysis, tumor immunohistochemistry (e.g., Her-2/neu, estrogen receptor/progesterone receptor), quantitative or semiquantitative, per specimen, each single antibody stain procedure; using computer-assisted technology |
| 88367 |
Morphometric analysis, in situ hybridization (quantitative or semi-quantitative), using computer-assisted technology, per specimen; initial single probe stain procedure |
| 88368 |
Morphometric analysis, in situ hybridization (quantitative or semi-quantitative), manual, per specimen; initial single probe stain procedure |
| 88381 |
Microdissection (i.e., sample preparation of microscopically identified target); manual |
| 0045U |
Oncology (breast ductal carcinoma in situ), mRNA, gene expression profiling by real-time RT-PCR of 12 genes (7 content and 5 housekeeping), utilizing formalin-fixed paraffin-embedded tissue, algorithm reported as recurrence score |
| 0067U |
Oncology (breast), immunohistochemistry, protein expression profiling of 4 biomarkers (matrix metalloproteinase-1 [MMP-1], carcinoembryonic antigen-related cell adhesion molecule 6 [CEACAM6], hyaluronoglucosaminidase [HYAL1], highly expressed in cancer protein [HEC1]), formalin-fixed paraffin-embedded precancerous breast tissue, algorithm reported as carcinoma risk score Proprietary test: BBDRisk Dx™ Lab/manufacturer: Silbiotech, Inc. |
| 0153U |
Oncology (breast), mRNA, gene expression profiling by next-generation sequencing of 101 genes, utilizing formalin-fixed paraffin-embedded tissue, algorithm reported as a triple negative breast cancer clinical subtype(s) with information on immune cell involvement |
| 0295U |
Oncology (breast ductal carcinoma in situ), protein expression profiling by immunohistochemistry of 7 proteins (COX2, FOXA1, HER2, Ki-67, p16, PR, SIAH2), with 4 clinicopathologic factors (size, age, margin status, palpability), utilizing formalin-fixed paraffin-embedded (FFPE) tissue, algorithm reported as a recurrence risk score Proprietary test: DCISionRT® Lab/Manufacturer: PreludeDx™/Prelude Corporation |
| S3854 |
Gene expression profiling panel for use in the management of breast cancer treatment |
Procedure and diagnosis codes on Medical Policy documents are included only as a general reference tool for each policy. They may not be all-inclusive.
This medical policy was developed through consideration of peer-reviewed medical literature generally recognized by the relevant medical community, U.S. FDA approval status, nationally accepted standards of medical practice and accepted standards of medical practice in this community, and other nonaffiliated technology evaluation centers, reference to federal regulations, other plan medical policies, and accredited national guidelines.
"Current Procedural Terminology © American Medical Association. All Rights Reserved"
History From 2014 Forward
| 04/08/2025 | Annual review, no change to policy intent. Updating description, rationale, guidelines/recommendations, and references. |
| 05/06/2024 | Annual review, policy updated for clarity and consistency. Criteria #2 has been expanded to include specifics about the tumor type and size Also updating rationale and references. |
| 04/19/2023 | Annual review, policy updated for clarity and consistency and addition of test brand names. Also updating description, policy, title, rationale, references, notes and removing 84999 from coding. |
| 04/26/2022 |
Annual review, updating policy for clarity in criteria 8 and 9. Adding Criteria 11 regarding DCISionRT and BBDRiskDX. Also updating rationale and references. Adding table of terminology. |
| 12/8/2021 |
Updating policy with 2022 coding. Adding code 81523. No other change made. |
| 04/19/2021 |
Annual review, updating policy to expand coverage ( see new coverage criteria #4. Also updating description, rationale and references. |
| 04/20/2020 |
Annual review, updating testing covered per NCCN guidelines. |
| 01/02/2020 |
Interim review, rewriting policy criteria for clarity. Also updating coding. |
| 07/12/2019 |
Annual review, updating coding and policy. Policy updates are in line with NCCN recommendations and relate to tumor site and type, potential treatment, staging and timing of diagnosis. Updating verbiage related to testing for males for clarity. |
| 12/21/2018 |
Updating with 2019 codes. |
| 07/24/2018 |
Annual review, reformatting entire policy. Expanding medical necessity criteria to allow Mammaprint testing for some indications. |
| 12/7/2017 |
Updating policy with 2018 coding. No other changes. |
| 07/19/2017 |
Annual review, updating policy criteria related to DX 21 gene expression, otherwise, no change to policy intent. |
| 04/25/2017 |
Updated category to Laboratory. No other changes |
| 12/06/2016 |
Interim review, adding medical necessity for Prosigna and EndoPredict testing. Updating background, description, guidelines, regulatory status, rationale and references. |
| 02/01/2016 |
Annual review, no change to policy intent. Updated background, description, guidelines, rationale and references. |
| 02/16/2015 |
Interim review, added additional criteria for medical necessity regarding the histology of the tumor, added additional investigational tests, added that gene expression profiling as a technique of managing the treatment of ductal cardinoma in situ is investigational and that repeat gene expression profiling is investigational. |
| 01/28/2015 |
Annual review, the following added to policy:The use of other gene expression assays, MammaPrint® 70-gene signature, Mammostrat® Breast Cancer Test, the Breast Cancer Index SM, BreastOncPx™, NexCourse® Breast IHC4, Prosigna™ BreastPRS™, and EndoPredict™ for any indication is considered investigational. |
| 01/30/2014 |
Annual Review |