Molecular Profiling for Cancers of Unknown Primary Origin - CAM 298
Description
Cancers of unknown primary origin (CUPs) are defined as the four to five percent of invasive cancers for which no primary site can be identified despite an extensive diagnostic work-up (Fizazi et al., 2015; Varadhachary & Raber, 2014). The American Cancer Society (ACS) estimates that about 34,950 cases of cancer of unknown primary will be diagnosed in 2024 in the United States (ACS, 2024). CUPs are generally considered to represent metastases and are associated with a very poor prognosis (Vikeså et al., 2015).
Gene expression assays measure the number of specific messenger RNAs (mRNAs) being transcribed to assess the genes that are active in a particular cell or tissue. Analyses of gene expression can be clinically useful for disease classification, diagnosis, prognosis, and tailoring treatment to underlying genetic determinants of pharmacologic response (Steiling, 2023).
For guidance concerning Tumor Mutational Burden Testing (TMB) and/or Microsatellite instability (MSI) analysis, please refer to CAM 342-Microsatellite Instability and Tumor Mutational Burden Testing policy.
Policy
Application of coverage criteria is dependent upon an individual’s benefit coverage at the time of the request.
The following does not meet coverage criteria due to a lack of available published scientific literature confirming that the test(s) is/are required and beneficial for the diagnosis and treatment of an individual’s illness.
- To evaluate the site of origin of a tumor of unknown primary, to distinguish a primary from a metastatic tumor, or to guide site-specific therapy, molecular cancer classifiers and/or gene expression profiling assays is considered NOT MEDICALLY NECESSARY.
Table of Terminology
| Term |
Definition |
| ACS |
American Cancer Society |
| ACUP |
Adeno carcinomas of unknown primary site |
| ARID1A |
AT-rich interactive domain-containing protein 1A |
| CDKN2A |
Cyclin-dependent kinase inhibitor 2a |
| CUP |
Cancers of unknown primary origin |
| ESMO |
European Society of Medical Oncology |
| FFPE |
Formalin-fixed paraffin-embedded |
| Gas |
Genomic alterations |
| GEP |
Gene expression profiling |
| ICI |
Immune checkpoint inhibitor |
| IHC |
Immunohistochemistry |
| KRAS |
Kirsten rat sarcoma viral oncogene homolog |
| MAKP |
Mitogen-Activated Protein Kinase |
| MDM2 |
Mouse double minute 2 homolog |
| MSI |
Microsatellite instability |
| mRNA |
Messenger ribonucleic acid |
| NCCN |
National Comprehensive Cancer Network |
| NCI |
National Cancer Institute |
| NICE |
National Institute for Health and Clinical Excellence |
| NSCLC |
Non-small cell lung cancer |
| PD-L1 |
Programmed death-ligand 1 |
| RTK |
Receptor tyrosine kinase |
| RT-PCR |
Reverse transcription polymerase chain reaction |
| SMAD4 |
SMAD family member 4 |
| TML |
Total mutational load |
| TNBC |
Triple-negative breast cancer |
| TP53 |
Tumor protein p53 |
Rationale
Cancers of unknown primary origin (CUPs) typically present with symptoms attributable to metastases where subsequent work-up fails to identify the primary site (J. Hainsworth & F. Greco, 2023). Given their rapid progression and dissemination, it was assumed that regardless of the site of origin, the tumors in unknown primary cancers shared biologic properties common to their pathogenesis and that identification of the exact tissue of origin would not have a substantial effect on therapeutic approaches or survival. However, biologic events that allow development of metastases without a discernable tumor at the primary site have not yet been determined (Varadhachary & Raber, 2014).
Accurate prediction of the tissue of origin using immunohistochemical staining and/or gene expression profiling is now possible in certain CUP cases. Appropriate classification, based upon all available evidence, is essential to identify patients for whom a specific treatment may be particularly useful and site-specific therapy based on these predictions is replacing empiric chemotherapy as the new treatment standard (J. Hainsworth & F. Greco, 2023). Tumors in unknown primary cancer despite different degrees of loss of differentiation retain the signature of their primary origin, even after metastasis (Fizazi et al., 2015).
Presently, patients are initially placed into one of four categories (adenocarcinoma, squamous cell carcinoma, neuroendocrine carcinoma, poorly differentiated) based upon the light microscopic examination of the initial biopsy. This classification is then used to guide further evaluation as indicated below (J. D. Hainsworth & F. A. Greco, 2023):
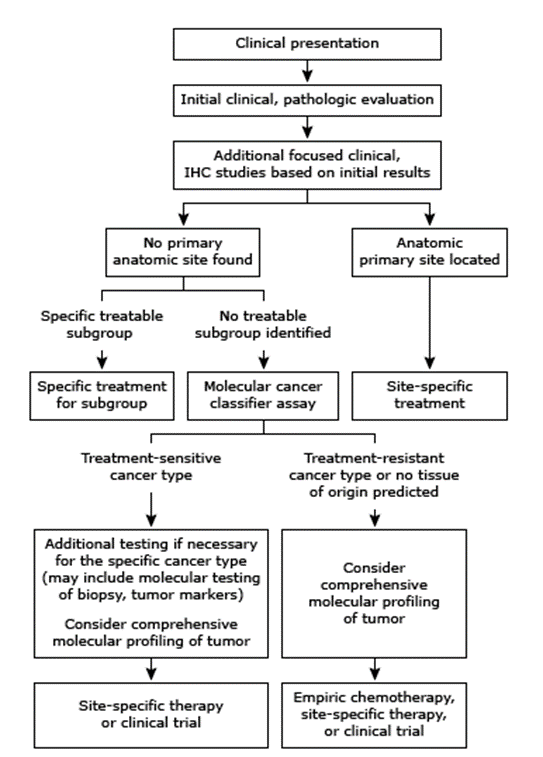
AAlthough the true tissue of origin may not be identifiable in any given case, recent evidence suggests it may be useful to provide “site-specific” treatment based on tumor type (J. Hainsworth & F. Greco, 2023). Certain characteristics of a tumor, such as its histology, may indicate more responsive cases and as such, may warrant specific treatments. For example, an individual with peritoneal carcinomatosis may benefit from therapies that are effective against advanced epithelial ovarian cancer, as the histology in both types may be similar. However, although several of these subgroups have been identified, most patients will not fall in these groups; approximately 70% of cancers of unknown primary are classified as adenocarcinomas, and 80-85% of these adenocarcinomas are not yet classified into these subgroups. In the absence of a targeted therapy, “empiric” chemotherapy with efficacy against a broad swath of cancer types is usually provided (J. Hainsworth & F. Greco, 2023; Hainsworth, 2024).
Proprietary Testing
Several proprietary tests exist for the assessment of the origin of cancer. One of these tests is “Tissue of Origin” from Cancer Genetics Inc. This test assesses the expression level of over 2000 genes and reports the likeliest tissue of origin from the most 15 common tumor types (“breast, non-small cell lung, pancreas, gastric, colorectal, liver, bladder, kidney, thyroid, non-Hodgkin lymphoma, melanoma, ovarian, sarcoma, testicular germ cell, and prostate”). An RNA “profile” is generated from the expression levels and compared to tissue profiles representative of the 15 tumors (Cancer Genetics Inc., 2018).
Tissue of Origin was validated by Pillai et al. (2011) where they created microarray data files for 462 “metastatic, poorly differentiated, or undifferentiated FFPE tumor specimens, all of which had a reference diagnosis” and analyzed these files with the Tissue of Origin model. Overall agreement with the reference diagnosis was 89%, and an average of 12 tissues could be ruled out with >99% probability (Pillai et al., 2011). Nystrom et al. (2012) examined the utility of this test by sending a survey to 65 physicians overseeing 107 patients. They found that, with the gene expression profile results, the diagnosis was changed for 50% of patients and management was changed for 65% of patients (Nystrom et al., 2012).
Hologic (2024) released CancerTYPE ID used to identify tumor origin in metastatic cancers. CancerTYPE ID is a gene expression assay that uses “real-time RT-PCR to measure the expression of 92-genes in the patient's tumor and classifies the tumor by matching the gene expression pattern to a database of over 2,000 known tumor types and subtypes. CancerTYPE ID can differentiate between 50 different tumor types and subtypes, covering >95% of all solid tumors based on incidence” (Hologic, 2024). CancerTYPE ID has been validated with 87% accuracy, 98% tumor type identified, and 37% improved survival.
Other than proprietary tests, The Jackson Laboratory developed CUP-AI-Dx, an RNA-based classifier, that uses RNA sequencing data from 817 genes to determine the metastatic cancer's primary tissue of origin and identify a tumor's molecular subtype. This machine was trained with the transcriptional profiles of 18,217 primary tumors and 32 cancer types. CUP-AI-Dx may be an important tool to help guide therapies for those who are limited to generalized treatment approaches (Zhao et al., 2020).
Analytical Validity
Kerr et al. (2012) conducted a large multi-institution validation study to examine the performance of a 92-gene molecular cancer classifier. The assay showed overall sensitivities of 87% for tumor type and 82% for subtype. No decrease in comparative performance was observed when metastatic tumors, high-grade tumors or cases with limited tissue were analyzed. The authors concluded that the assay showed strong performance for accurate molecular classification for various tumor histologies. They further state that “results support potential use of the assay as a standardized molecular adjunct to routine clinicopathologic evaluation for tumor classification and primary site diagnosis” (Kerr et al., 2012).
Handorf et al. (2013) published the results of a prospectively conducted, blinded, multicenter study that compared the diagnostic accuracy of gene expression profiling (GEP) with IHC in identifying the primary site of metastatic tumors with known primaries. Overall, GEP accurately identified 89% of specimens, compared with 83% accuracy using IHC. In the subset of 33 poorly differentiated and undifferentiated carcinomas, GEP had higher accuracy (91%) compared to IHC (71%). The authors concluded that GEP “was significantly more accurate than IHC when used to identify the primary site of metastatic tumors” (Handorf et al., 2013).
In a similar study design, Handorf et al. (2013) compared the diagnostic accuracy of IHC analysis versus molecular classification using a 92-gene RT-PCR assay for determination of the primary tumor site. The authors reported 79% accuracy for gene expression profiling compared with 69% for immunohistochemistry. The authors concluded that the results “demonstrate superior accuracy with the 92-gene assay versus standard-of-care IHC analysis and strongly support the diagnostic utility of molecular classification in difficult-to-diagnose metastatic cancer” (Weiss et al., 2013).
Loffler et al. (2016) performed next-generation sequencing (of 50 genes) on 55 patients with adenocarcinoma or undifferentiated carcinoma. 46 cases harbored tumor-specific mutations and other alterations. TP53, KRAS, CDKN2A and SMAD4 were the most mutated genes, and 8 cases were identified as having targetable mutations by currently approved drugs. The authors concluded that mutations of relevant driver genes were present in “vast majority” of CUP tumors and that these genes may carry impact on prognosis and targeted therapy” (Loffler et al., 2016).
Santos et al. (2017) aimed to develop and validate a gene-expression classifier to identify potential primary sites for metastatic cancers more accurately. “The gene-expression classifier correctly identified, by a cross-validation, 86.6% of the expected cancer superclasses of 4429 samples from the RefDB, with a specificity of 99.43%. Next, the performance of the algorithm for classifying the validation set of metastatic FFPE samples was 83.81%, with 99.04% specificity. The overall reproducibility of our gene-expression-classifier system was 97.22% of precision, with a coefficient of variation for inter-assays and intra-assays and intra-lots < 4.1%” (Santos et al., 2017).
In a study by Zhao et al. (2020),the CUP-AI-DX was tested on 394 metastatic samples of unknown primary origin. The machine correctly identified the tissue of origin 96.7% of the time. The authors also compared the classification accuracy to the CancerTypeID GEP test. While the accuracy of CUP-AI-DX was 98.54% in cross-validation, while CancerType ID was 87% in cross-validation. The authors conclude that "The CUP-AI-Dx predicts tumour primary site and molecular subtype with high accuracy and therefore can be used to assist the diagnostic work-up of cancers of unknown primary origin” (Zhao et al., 2020).
Raghav et al. (2020) studied the use of CancerType ID GEP assay to identify cancers of unknown primary for immune checkpoint inhibitor (ICI) therapy. A total of 9,250 cases were studied, and the assay found that non-small cell lung cancer (NSCLC) accounted for 33% of the molecular diagnosis. The assay also frequently recognized urothelial carcinomas, gastric cancer, and head and neck squamous cell carcinoma, all of which could be treated with ICI therapy. This assay identified 40% of cases for which an FDA-approved ICI was available. Currently, ICIs are only indicated for CUP on rare occurrences; therefore, identifying ICI-eligible CUP patients with this assay is an important step towards improving treatments (Raghav et al., 2020).
Sun et al. (2022) studied the diagnostic utility of a 90-gene expression test for tumor classification. 1,417 samples were analyzed using the 90-gene expression test and the results were compared to histopathological diagnosis. Overall, the 90-gene expression test reached an accuracy of 94.4%. "Among different tumor types, sensitivities ranged from 74.2% (head & neck tumor) to 100% (adrenal carcinoma, mesothelioma, and prostate cancer). Sensitivities for the most prevalent cancers of lung, breast, colorectum, and gastroesophagus are 95.0%, 98.4%, 93.9%, and 90.6%, respectively. Moreover, specificities for all 21 tumor types are greater than 99%” (Sun et al., 2022). The authors conclude that this 90-gene expression test can be used as an adjunct for tumor classification in clinical practice.
Clinical Utility and Validity
Several studies have investigated the validity and diagnostic utility of gene expression profiling in addition to or in place of standard immunohistochemistry in the diagnosis and management of CUP.
Hainsworth et al. (2013) conducted a prospective trial testing the tumor biopsy specimens from previously untreated patients with CUP with a 92-gene reverse transcriptase polymerase chain reaction cancer classification assay. Molecular tumor profiling correctly identified tissue of origin in 85% of carcinomas of known primary origin. The study showed that molecular tumor profiling predicted a tissue of origin in 247 of 252 (98%) patients with CUP. The authors concluded that “molecular tumor profiling contributes to the management of patients with CUP and should be a part of their standard evaluation” (Hainsworth et al., 2013).
Greco et al. (2013) demonstrated that 18 of 24 patients (75%) with latent primaries discovered months to years later were predicted by molecular tumor profiling. The authors concluded that molecular tumor profiling “complements standard pathologic evaluation in determining the tissue of origin in patients with CUP, particularly when IHC is inconclusive” (Greco et al., 2013).
Oien and Dennis (2012) concluded that “in already well worked-up poorly differentiated and/or metastatic tumours, including CUP, molecular profiling performs well, with sensitivities of 72% – 95% and may outperform optimal IHC by 10%–20%” (Oien & Dennis, 2012). The authors conclude that molecular profiling could thus contribute to diagnosis of poorly differentiated and/or metastatic tumors.
Ross et al. (2015) conducted comprehensive genomic profiling on 200 CUP formalin-fixed paraffin-embedded specimens (mean, 756× coverage) using the hybrid-capture-based FoundationOne assay for presence of targetable genomic alterations (GAs) in CUP and responses to targeted therapies. They concluded that “Almost all CUP samples harbored at least 1 clinically relevant GA with potential to influence and personalize therapy. The ACUP [adenocarcinomas of unknown primary site] tumors were more frequently driven by GAs in the highly druggable RTK/Ras/mitogen-activated protein kinase (MAPK) signaling pathway than the non-ACUP tumors. Comprehensive genomic profiling can identify novel treatment paradigms to address the limited options and poor prognoses of patients with CUP” (Ross et al., 2015).
Groschel et al. (2016) investigated if their results from a difficult case could be extrapolated. The authors described an advanced-stage malignancy that mimicked a poorly differentiated soft-tissue sarcoma and did not respond to multiagent chemotherapy. Despite molecular profiling and histopathology analysis, the tissue of origin was not identified. However, the authors believed that immune checkpoint inhibitor therapy was warranted, and several differential diagnoses were theorized, including triple-negative breast cancer (TNBC). The authors assessed 157 TNBC cases from the Cancer Genome Atlas and found PD-L1 copy number gains (leading to excess PD-L1 mRNA expression) in 24% of cases. The authors concluded that their results “illustrate the impact of multidimensional tumor profiling in cases with nondescript histology and immunophenotype, show the predictive potential of PDL1 amplification for immune checkpoint inhibitors (ICIs), and suggest a targeted therapeutic strategy in Chromosome 9p24.1/PDL1-amplified cancers” (Groschel et al., 2016).
Zehir et al. (2017) attempted to characterize the mutational landscape of metastatic cancer. 10,945 tumor samples from 10,336 patients were included. Tumors were sequenced with two panels, one of 341 genes, and another of 410 genes (with all 341 genes from the former panel included). Tumors were sequenced to an average of 718x coverage. Non-small cell lung cancer was the most common, with 1,563 patients, followed by breast carcinoma at 1237 patients and colorectal cancer at 978 patients. Cancers of unknown primary comprised of a total of 160 patients. Overall, the authors identified 36.7% of patients as having actionable mutations (n = 3,792). Gastrointestinal stromal tumors (76%), thyroid cancer (60%) and breast cancer (57%) were found to have the highest proportion of actionable mutations. However, the highest standard of actionable mutation met for cancers of unknown primary was “level 2B”, or “standard of care biomarker for an FDA-approved drug in another indication”. The authors concluded that their data “demonstrate [d] the feasibility and utility of large-scale prospective clinical sequencing of matched tumor-normal pairs to guide clinical management (Zehir et al., 2017).
Varghese et al. (2021) aimed to provide a “clinical and pathologic” description of patients with cancers of unknown primary. A total of 150 patients had targeted next-generation sequencing performed. There were 45 patients identified to have “potentially actionable” mutations, and 15 patients received targeted therapies. The authors remarked that CUP patients may benefit from targeted therapies (Varghese et al., 2021).
Gatalica et al. (2018) attempted to identify predictive biomarkers for immune checkpoint blockade therapy in cancers of unknown primary (CUP). A total of 389 cases were analyzed for 592 mutations and 52 gene fusions through next-generation sequencing. Microsatellite instability (MSI), total mutational load (TML), and PD-L1 expression were all evaluated. The authors identified “high” TML in 11.8% of tumors, high MSI in 7 tumors, and PD-L1 expression in 80 (of 362 tested cases) tumors. Other predictive biomarkers such as MDM2 gene amplification were identified. TP53 gene mutations were found in 54% of cases, followed by KRAS (22%) and ARID1A (13%). Overall, the authors identified 28% of CUP cases as carrying a predictive biomarker for immune checkpoint blockade therapy (Gatalica et al., 2018).
Clynick et al. (2018) attempted to identify actionable mutations in cancers of unknown primary. There were 21 cases included, and two gene panels were used to evaluate variants in 76 cancer-related genes. The authors found variants in 17 of 21 cases, with 11 considered “potentially actionable.” The most common variants detected were TP53 (47%), KRAS (12%), MET (12%) and MYC (12%). The authors also remarked that CUP adenocarcinomas and poorly differentiated carcinomas tended to harbor gene mutations involved in signal transduction pathways (eight of eleven cases harboring mutations such as BRAF, HRAS, and KRAS), whereas squamous cell carcinomas tended to harbor mutations in genes involved in cell cycle control and DNA repair genes (all eight cases harboring mutations such as TP53, MLH1, and CDKN2A). Overall, the authors identified mutations in “biologically relevant” genes in the “vast majority” of CUP tumors, noting that half provided a “potentially novel treatment not generally considered in CUP” (Clynick et al., 2018).
Hayashi et al. (2019) compared two treatments for cancers of unknown primary site. Empirical chemotherapy was compared against site-directed therapy (directed by comprehensive microarray-based gene expression profiling). Efficacy analysis was performed for 50 patients in the site-specific arm and 51 patients in the empirical chemotherapy arm. One-year survival rate was found to be 44% for site-specific treatment and 54.9% for empirical treatment (p = .264). Median overall and progression-free survival was found to be 9.8 months and 5.1 months respectively for site-specific treatment and 12.5 months and 4.8 months respectively for empirical treatment (p=.896 and .550, respectively). Overall, the authors concluded that “Site-specific treatment that was based on microarray profiling did not result in a significant improvement in one-year survival compared with empirical PC [treatment]” (Hayashi et al., 2019).
Fizazi et al. (2019) evaluated the utility of “tailored treatment” for cancers of unknown primary. 243 patients with cancers of unknown primary were included and were randomized to Arm A (“Cisplatin 100 mg/m2 d1þ Gemcitabine 1250 mg/m2, day one and eight, q3w”, empiric treatment) or Arm B (“gene expression test followed by a la carte treatment according to the suspected primary”, tailored). The gene expression tests used were Pathwork’s Tissue of Origin (n = 21) or CancerTYPE (n = 222), and the primary endpoint was defined as progression-free survival at a hazard ratio of 0.625. The four most common tissues of origin were “pancreatico-biliary cancer (19%), squamous cell carcinoma (eleven percent, kidney cancer (eight percent) and lung cancer (eight percent)”. A total of 91 out of the 123 patients in Arm B were given tailored treatment. Progression-free survival in both arms were similar (both by central and local review), overall survival was similar in both arms (hazard ratio = 0.92). Overall, the authors concluded that “using a molecular test followed by tailored systemic treatment did not improve outcomes of pts [patients] with CUP” (Fizazi et al., 2019).
Cobain et al. (2021) studied which patients have the greatest clinical benefit from NGS profiling. NGS was performed in 1,015 patients and clinically actionable genomic alterations were found in 817 patients (80.5%). Of the 817 patients, 132 (16.2%) received sequencing-directed therapy, and 49 had clinical benefit (37.1%). "For 55 patients with carcinoma of unknown primary origin, NGS identified the primary site in 28 (50.9%), and sequencing-directed therapy in 13 patients resulted in clinical benefit in 7 instances (53.8%), including 5 exceptional responses" (Cobain et al., 2021). The authors conclude directed germline testing and genomic profiling should be used as a standard of care for patients with cancer of unknown origin.
Saeed et al. (2022) studied the utility and impact of genomic profiling to determine tissue origin of CUPs. The study included tissue or cytology specimens from 22 CUPs, 15 of which were adequate for analysis. “Primary tumor site was suggested in 12 cases (80%), whereas it remained indeterminate in 3 (20%).” The doctors concluded that “genomic profiling helped confirm the original diagnosis and suggested primary sites in two third of our cases” (Saeed et al., 2022).
Posner et al. (2023) compared gene expression profiling (GEP) and DNA sequencing as tools for predicting primary tissue of origin in CUP. The study included 215 CUP patients, 82% of whom received both tests. Based on retrospective clinicopathological data, 77%, of cases had insufficient evidence to support a tissue of origin diagnosis. After DNA sequencing, “mutations and mutational signatures provided additional diagnostic evidence in 31% of cases.” Alternatively, “GEP classification was useful in only 13% of cases and oncoviral detection in 4%.” The authors conclude that DNA mutation profiling was “the more diagnostically informative assay” compared to GEP (Posner et al., 2023).
National Comprehensive Cancer Network (NCCN)
The NCCN lists two primary applications of molecular profiling in management of CUP; using gene expression profiling and molecular cancer classifier assays to determine tissue of origin for site-specific therapy, and identifying actionable mutations for targeted therapy (NCCN, 2024).
The 2023 NCCN guidelines for the workup of an occult primary malignancy address the use of molecular methods in the classification of tumors. The guidelines state “Gene sequencing to predict tissue of origin is not recommended.” The guidelines also state that “molecular profiling of tumor tissue using NGS or other technique to identify gene fusions can be considered after an initial determination of histology has been made.” Further, the guidelines note that “While there may be a diagnostic benefit to GEP, a clinical benefit has not been demonstrated.” The guidelines further recommend that “until more robust outcomes and comparative effectiveness data are available, pathologists and oncologists must collaborate on the judicious use of IHC and GEP on a case-by-case basis, with the best possible individualized patient outcome in mind” (NCCN, 2024).
Overall, the NCCN states that “the clinical benefit of using molecular profiling to guide treatment decisions in CUP remains to be determined.” The NCCN also states that “currently, there is no evidence of improved outcomes with the use of site-specific therapy guided by molecular testing results in patients with CUP” (NCCN, 2024).
National Institute for Health and Clinical Excellence (NICE)
A 2010 clinical guideline from NICE, which was reaffirmed in 2017, recommended against the use of gene expression-based profiling to identify primary tumors in patients with provisional CUPs. The guideline also states, “do not use gene-expression-based profiling when deciding which treatment to offer patients with confirmed CUP” (NICE, 2010).
In the April 2023 update of the metastatic malignant disease of unknown primary origin in adults: diagnosis and management clinical guideline, NICE withdrew recommendations on gene-expression-based profiling (NICE, 2010).
European Society of Medical Oncology (ESMO)
In 2023, EMSO updated their clinical practice guidelines for the diagnosis, treatment, and follow-up of cancer of unknown primary. The guideline states that “The clinical utility of gene expression profiling to help elucidate the likely primary is not currently supported by high-level evidence. Consequently, it is not generally recommended outside of clinical research.” Additionally, “there is currently no high-level evidence that gene expression profiling-directed therapy leads to an improvement in patient outcomes. Consequently, such strategies are not recommended outside of clinical trials” (Krämer et al., 2023).
National Cancer Institute (NCI)
The NCI acknowledges the possible utility of gene expression profiling and next generation sequencing to identify a potential site of origin in patients with CUP (NCI, 2023).
American Cancer Society (ACS)
The American Cancer Society acknowledges the possible use of gene expression profiling and molecule genetic testing for the diagnosis of cancers of unknown primary. However, they note that molecular genetic testing “is not needed in most cases, but it’s sometimes helpful in classifying some cancers when other tests have not provided clues regarding their origin.” Regarding gene expression profiling, the ACS states that “these tests can sometimes help your doctor discover where the cancer may have started, but so far, they haven’t been linked to better outcomes in patients” (ACS, 2018; Losa et al., 2018).
References
- ACS. (2018). Tests for Cancer of Unknown Primary. https://www.cancer.org/cancer/cancer-unknown-primary/detection-diagnosis-staging/how-diagnosed.html
- ACS. (2024, January 17, 2024). Key Statistics for Cancers of Unknown Primary. https://www.cancer.org/cancer/cancer-unknown-primary/about/key-statistics.html
- Cancer Genetics Inc. (2018). Cancer Genetics’ Unique Tissue Of Origin Test (TOO®) Receives Special FDA 510(K) Clearance. https://www.globenewswire.com/news-release/2018/04/16/1471915/0/en/Cancer-Genetics-Unique-Tissue-of-Origin-Test-TOO-Receives-Special-FDA-510-k-Clearance.html
- Clynick, B., Dessauvagie, B., Sterrett, G., Harvey, N. T., Allcock, R. J. N., Saunders, C., Erber, W., & Meehan, K. (2018). Genetic characterisation of molecular targets in carcinoma of unknown primary. J Transl Med, 16(1), 185. https://doi.org/10.1186/s12967-018-1564-x
- Cobain, E. F., Wu, Y.-M., Vats, P., Chugh, R., Worden, F., Smith, D. C., Schuetze, S. M., Zalupski, M. M., Sahai, V., Alva, A., Schott, A. F., Caram, M. E. V., Hayes, D. F., Stoffel, E. M., Jacobs, M. F., Kumar-Sinha, C., Cao, X., Wang, R., Lucas, D., . . . Chinnaiyan, A. M. (2021). Assessment of Clinical Benefit of Integrative Genomic Profiling in Advanced Solid Tumors. JAMA Oncol, 7(4), 525-533. https://doi.org/10.1001/jamaoncol.2020.7987
- Fizazi, K., Greco, F. A., Pavlidis, N., Daugaard, G., Oien, K., & Pentheroudakis, G. (2015). Cancers of unknown primary site: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol, 26 Suppl 5, v133-138. https://doi.org/10.1093/annonc/mdv305
- Fizazi, K., Maillard, A., Penel, N., Baciarello, G., Allouache, D., Daugaard, G., Van de Wouw, A., Soler, G., Vauleon, E., Chaigneau, L., Janssen, R., Losa Gaspa, F., Morales Barrera, R., Balana, C., Tosi, D., Chauffert, B., Schnabel, C. A., Martineau, G., Culine, S., & Borget, I. (2019). LBA15_PR - A phase III trial of empiric chemotherapy with cisplatin and gemcitabine or systemic treatment tailored by molecular gene expression analysis in patients with carcinomas of an unknown primary (CUP) site (GEFCAPI 04). Annals of Oncology, 30, v851. https://doi.org/10.1093/annonc/mdz394
- Gatalica, Z., Xiu, J., Swensen, J., & Vranic, S. (2018). Comprehensive analysis of cancers of unknown primary for the biomarkers of response to immune checkpoint blockade therapy. Eur J Cancer, 94, 179-186. https://doi.org/10.1016/j.ejca.2018.02.021
- Greco, F. A., Lennington, W. J., Spigel, D. R., & Hainsworth, J. D. (2013). Molecular profiling diagnosis in unknown primary cancer: accuracy and ability to complement standard pathology. J Natl Cancer Inst, 105(11), 782-790. https://doi.org/10.1093/jnci/djt099
- Groschel, S., Bommer, M., Hutter, B., Budczies, J., Bonekamp, D., Heining, C., Horak, P., Frohlich, M., Uhrig, S., Hubschmann, D., Georg, C., Richter, D., Pfarr, N., Pfutze, K., Wolf, S., Schirmacher, P., Jager, D., von Kalle, C., Brors, B., . . . Frohling, S. (2016). Integration of genomics and histology revises diagnosis and enables effective therapy of refractory cancer of unknown primary with PDL1 amplification. Cold Spring Harb Mol Case Stud, 2(6), a001180. https://doi.org/10.1101/mcs.a001180
- Hainsworth, J., & Greco, F. (2023, 12/15/2023). Overview of the classification and management of cancers of unknown primary site. https://www.uptodate.com/contents/overview-of-the-classification-and-management-of-cancers-of-unknown-primary-site
- Hainsworth, J. D., & Greco, F. A. (2023, 12/15/2023). Treatment of patients with cancer of unknown primary site. https://www.uptodate.com/contents/overview-of-the-classification-and-management-of-cancers-of-unknown-primary-site#topicGraphics
- Hainsworth, J. D., Greco, Anthony. (2024, 01/31/2024). Adenocarcinoma of unknown primary site. https://www.uptodate.com/contents/adenocarcinoma-of-unknown-primary-site
- Hainsworth, J. D., Rubin, M. S., Spigel, D. R., Boccia, R. V., Raby, S., Quinn, R., & Greco, F. A. (2013). Molecular gene expression profiling to predict the tissue of origin and direct site-specific therapy in patients with carcinoma of unknown primary site: a prospective trial of the Sarah Cannon research institute. J Clin Oncol, 31(2), 217-223. https://doi.org/10.1200/jco.2012.43.3755
- Handorf, C. R., Kulkarni, A., Grenert, J. P., Weiss, L. M., Rogers, W. M., Kim, O. S., Monzon, F. A., Halks-Miller, M., Anderson, G. G., Walker, M. G., Pillai, R., & Henner, W. D. (2013). A multicenter study directly comparing the diagnostic accuracy of gene expression profiling and immunohistochemistry for primary site identification in metastatic tumors. Am J Surg Pathol, 37(7), 1067-1075. https://doi.org/10.1097/PAS.0b013e31828309c4
- Hayashi, H., Kurata, T., Takiguchi, Y., Arai, M., Takeda, K., Akiyoshi, K., Matsumoto, K., Onoe, T., Mukai, H., Matsubara, N., Minami, H., Toyoda, M., Onozawa, Y., Ono, A., Fujita, Y., Sakai, K., Koh, Y., Takeuchi, A., Ohashi, Y., . . . Nakagawa, K. (2019). Randomized Phase II Trial Comparing Site-Specific Treatment Based on Gene Expression Profiling With Carboplatin and Paclitaxel for Patients With Cancer of Unknown Primary Site. J Clin Oncol, 37(7), 570-579. https://doi.org/10.1200/jco.18.00771
- Hologic. (2024). CancerTYPE ID. https://www.hologic.com/hologic-products/diagnostic-solutions/cancertype-id
- Kerr, S. E., Schnabel, C. A., Sullivan, P. S., Zhang, Y., Singh, V., Carey, B., Erlander, M. G., Highsmith, W. E., Dry, S. M., & Brachtel, E. F. (2012). Multisite validation study to determine performance characteristics of a 92-gene molecular cancer classifier. Clin Cancer Res, 18(14), 3952-3960. https://doi.org/10.1158/1078-0432.ccr-12-0920
- Krämer, A., Bochtler, T., Pauli, C., Baciarello, G., Delorme, S., Hemminki, K., Mileshkin, L., Moch, H., Oien, K., Olivier, T., Patrikidou, A., Wasan, H., Zarkavelis, G., Pentheroudakis, G., & Fizazi, K. (2023). Cancer of unknown primary: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol, 34(3), 228-246. https://doi.org/10.1016/j.annonc.2022.11.013
- Loffler, H., Pfarr, N., Kriegsmann, M., Endris, V., Hielscher, T., Lohneis, P., Folprecht, G., Stenzinger, A., Dietel, M., Weichert, W., & Kramer, A. (2016). Molecular driver alterations and their clinical relevance in cancer of unknown primary site. Oncotarget, 7(28), 44322-44329. https://doi.org/10.18632/oncotarget.10035
- Losa, F., Soler, G., Casado, A., Estival, A., Fernández, I., Giménez, S., Longo, F., Pazo-Cid, R., Salgado, J., & Seguí, M. (2018). SEOM clinical guideline on unknown primary cancer (2017). Clin Transl Oncol, 20(1), 89-96. https://doi.org/10.1007/s12094-017-1807-y
- NCCN. (2024). Occult Primary (Cancer of Unknown Primary [CUP]) Version 1.2024. National Comprehensive Cancer Network. https://www.nccn.org/professionals/physician_gls/pdf/occult.pdf
- NCI. (2023, December 22, 2023). Carcinoma of Unknown Primary Treatment (PDQ®)–Health Professional Version. https://www.cancer.gov/types/unknown-primary/hp/unknown-primary-treatment-pdq#_10
- NICE. (2010, 04/26/2023). Metastatic malignant disease of unknown primary origin in adults: diagnosis and management | Guidance and guidelines | NICE. NICE. https://www.nice.org.uk/guidance/cg104
- Nystrom, S. J., Hornberger, J. C., Varadhachary, G. R., Hornberger, R. J., Gutierrez, H. R., Henner, D. W., Becker, S. H., Amin, M. B., & Walker, M. G. (2012). Clinical utility of gene-expression profiling for tumor-site origin in patients with metastatic or poorly differentiated cancer: impact on diagnosis, treatment, and survival. Oncotarget, 3(6), 620-628. https://doi.org/10.18632/oncotarget.521
- Oien, K. A., & Dennis, J. L. (2012). Diagnostic work-up of carcinoma of unknown primary: from immunohistochemistry to molecular profiling. In Ann Oncol (Vol. 23 Suppl 10, pp. x271-277). https://doi.org/10.1093/annonc/mds357
- Pillai, R., Deeter, R., Rigl, C. T., Nystrom, J. S., Miller, M. H., Buturovic, L., & Henner, W. D. (2011). Validation and reproducibility of a microarray-based gene expression test for tumor identification in formalin-fixed, paraffin-embedded specimens. J Mol Diagn, 13(1), 48-56. https://doi.org/10.1016/j.jmoldx.2010.11.001
- Posner, A., Prall, O. W., Sivakumaran, T., Etemadamoghadam, D., Thio, N., Pattison, A., Balachander, S., Fisher, K., Webb, S., Wood, C., DeFazio, A., Wilcken, N., Gao, B., Karapetis, C. S., Singh, M., Collins, I. M., Richardson, G., Steer, C., Warren, M., . . . Tothill, R. W. (2023). A comparison of DNA sequencing and gene expression profiling to assist tissue of origin diagnosis in cancer of unknown primary. J Pathol, 259(1), 81-92. https://doi.org/10.1002/path.6022
- Raghav, K., Overman, M., Poage, G. M., Soifer, H. S., Schnabel, C. A., & Varadhachary, G. R. (2020). Defining a Distinct Immunotherapy Eligible Subset of Patients with Cancer of Unknown Primary Using Gene Expression Profiling with the 92-Gene Assay. Oncologist, 25(11), e1807-e1811. https://doi.org/10.1634/theoncologist.2020-0234
- Ross, J. S., Wang, K., Gay, L., Otto, G. A., White, E., Iwanik, K., Palmer, G., Yelensky, R., Lipson, D. M., Chmielecki, J., Erlich, R. L., Rankin, A. N., Ali, S. M., Elvin, J. A., Morosini, D., Miller, V. A., & Stephens, P. J. (2015). Comprehensive Genomic Profiling of Carcinoma of Unknown Primary Site: New Routes to Targeted Therapies. JAMA Oncol, 1(1), 40-49. https://doi.org/10.1001/jamaoncol.2014.216
- Saeed, O. A. M., Armutlu, A., Cheng, L., Longe, H. O., & Saxena, R. (2022). Tumor Genomic Profiling to Determine Tissue Origin of Cancers of Unknown Primary: A Single Institute Experience With its Utility and Impact on Patient Management. Appl Immunohistochem Mol Morphol, 30(9), 592-599. https://doi.org/10.1097/pai.0000000000001057
- Santos, M. T. D., Souza, B. F., Carcano, F. M., Vidal, R. O., Scapulatempo-Neto, C., Viana, C. R., & Carvalho, A. L. (2017). An integrated tool for determining the primary origin site of metastatic tumours. J Clin Pathol. https://doi.org/10.1136/jclinpath-2017-204887
- Steiling, K., Christenson, Stephanie. (2023, 10/03/2023). Tools for genetics and genomics: Gene expression profiling. https://www.uptodate.com/contents/tools-for-genetics-and-genomics-gene-expression-profiling
- Sun, W., Wu, W., Wang, Q., Yao, Q., Feng, Q., Wang, Y., Sun, Y., Liu, Y., Lai, Q., Zhang, G., Qi, P., Sun, Y., Qian, C., Ren, W., Luo, Z., Chen, J., Wang, H., Xu, Q., Zhou, X., . . . Lin, D. (2022). Clinical validation of a 90-gene expression test for tumor tissue of origin diagnosis: a large-scale multicenter study of 1417 patients. Journal of Translational Medicine, 20(1), 114. https://doi.org/10.1186/s12967-022-03318-6
- Varadhachary, G. R., & Raber, M. N. (2014). Cancer of unknown primary site. N Engl J Med, 371(8), 757-765. https://doi.org/10.1056/NEJMra1303917
- Varghese, A. M., Arora, A., Capanu, M., Camacho, N., Won, H. H., Zehir, A., Gao, J., Chakravarty, D., Schultz, N., Klimstra, D. S., Ladanyi, M., Hyman, D. M., Solit, D. B., Berger, M. F., & Saltz, L. B. (2021). Clinical and molecular characterization of patients with cancer of unknown primary in the modern era. Ann Oncol, 28(12), 3015-3021. https://doi.org/10.1093/annonc/mdx545
- Vikeså, J., Møller, A. K. H., Kaczkowski, B., Borup, R., Winther, O., Henao, R., Krogh, A., Perell, K., Jensen, F., Daugaard, G., & Nielsen, F. C. (2015). Cancers of unknown primary origin (CUP) are characterized by chromosomal instability (CIN) compared to metastasis of know origin. BMC Cancer, 15. https://doi.org/10.1186/s12885-015-1128-x
- Weiss, L. M., Chu, P., Schroeder, B. E., Singh, V., Zhang, Y., Erlander, M. G., & Schnabel, C. A. (2013). Blinded comparator study of immunohistochemical analysis versus a 92-gene cancer classifier in the diagnosis of the primary site in metastatic tumors. J Mol Diagn, 15(2), 263-269. https://doi.org/10.1016/j.jmoldx.2012.10.001
- Zehir, A., Benayed, R., Shah, R. H., Syed, A., Middha, S., Kim, H. R., Srinivasan, P., Gao, J., Chakravarty, D., Devlin, S. M., Hellmann, M. D., Barron, D. A., Schram, A. M., Hameed, M., Dogan, S., Ross, D. S., Hechtman, J. F., DeLair, D. F., Yao, J., . . . Berger, M. F. (2017). Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med, 23(6), 703-713. https://doi.org/10.1038/nm.4333
- Zhao, Y., Pan, Z., Namburi, S., Pattison, A., Posner, A., Balachander, S., Paisie, C. A., Reddi, H. V., Rueter, J., Gill, A. J., Fox, S., Raghav, K. P. S., Flynn, W. F., Tothill, R. W., Li, S., Karuturi, R. K. M., & George, J. (2020). CUP-AI-Dx: A tool for inferring cancer tissue of origin and molecular subtype using RNA gene-expression data and artificial intelligence. EBioMedicine, 61. https://doi.org/10.1016/j.ebiom.2020.103030
Coding Section
| Code | Number |
Description |
| CPT | 81479 |
Unlisted molecular pathology procedure |
| 81504 |
Oncology (tissue of origin), microarray gene expression profiling of > 2000 genes, utilizing formalin-fixed paraffin-embedded tissue, algorithm reported as tissue similarity scores |
|
| 81540 |
Oncology (tumor of unknown origin), mRNA, gene expression profiling by real-time RT-PCR of 92 genes (87 content and 5 housekeeping) to classify tumor into main cancer type and subtype, utilizing formalin-fixed paraffin-embedded tissue, algorithm reported as a probability of a predicted main cancer type and subtype |
|
| 81599 |
Unlisted multianalyte assay with algorithmic analysis |
|
| ICD-10-CM (effective 10/01/15) | Investigational for all relevant diagnoses | |
| C79.9 | Secondary malignant neoplasm of unspecified site | |
| C80.0 | Disseminated malignant neoplasm, unspecified | |
| C80.1 | Malignant (primary) neoplasm, unspecified | |
| ICD-10-PCS (effective 10/01/15) | Not applicable. ICD-10-PCS codes are only used for inpatient services. here are no ICD procedure codes for laboratory tests. | |
| Type of Service | Pathology/Laboratory | |
| Secondary malPlace of service | Laboratory/Reference Laboratory |
Procedure and diagnosis codes on Medical Policy documents are included only as a general reference tool for each policy. They may not be all-inclusive.
This medical policy was developed through consideration of peer-reviewed medical literature generally recognized by the relevant medical community, U.S. FDA approval status, nationally accepted standards of medical practice and accepted standards of medical practice in this community and other nonaffiliated technology evaluation centers, reference to federal regulations, other plan medical policies, and accredited national guidelines.
"Current Procedural Terminology © American Medical Association. All Rights Reserved"
History From 2013 Forward
| 07/30/2024 | Annual Review, no change to policy intent. Updated description, rationale and references. |
| 07/11/2023 | Annual review, no change to policy intent. Policy updated for clarity and consistency. Also updating description, table of terminology, rationale and references. Updating coding verbiage. |
| 07/19/2022 |
Annual review, no change to policy intent. Updating description, rationale and references. |
| 07/20/2021 |
Annual review, no change to policy intent. Updating description, rationale and references. Removing regulatory status as that is included in the rationale. |
| 07/21/2020 |
Annual review, updating policy criteria for specificity. Also updating description, rationale, references and title. |
| 07/12/2019 |
Annual review, no change to policy intent. |
| 07/18/2018 |
Annual review, no change to policy intent. |
| 07/17/2017 |
Annual review, no change to policy intent. Updating rationale and references. |
| 04/25/2017 |
Updated category to Laboratory. No other changes |
| 02/01/2017 |
Annual review, no change to policy intent. |
| 02/03/2016 |
Annual review, no change to policy intent. Updating background, description, guidelines, rationale and references. |
| 12/1/2015 |
Update CPT codes with 2016 codes. No change to intent of policy. |
| 02/25/2015 |
Annual review, no change to policy intent. Updated title, background, description, regulatory status, references and rationale. Added guidlines and coding. |
| 02/11/2014 |
Annual Review. Updated background, rationale and references. No change to policy intent. |